Difference between revisions of "Part:BBa K2549013"
(→Usage and Biology) |
|||
| Line 9: | Line 9: | ||
<!-- Add more about the biology of this part here --> | <!-- Add more about the biology of this part here --> | ||
| − | === | + | ===Applications=== |
| − | + | The main use of TEV protease<ref>https://en.wikipedia.org/wiki/TEV_protease</ref> is for removing affinity tags from purified recombinant fusion proteins. The reason for that is due to the high sequence specificity. This specificity allows for the controlled cleavage of proteins when the preference sequence is inserted into flexible loops. It also makes it relatively non-toxic in vivo as the recognized sequence scarcely occurs in proteins. | |
| + | |||
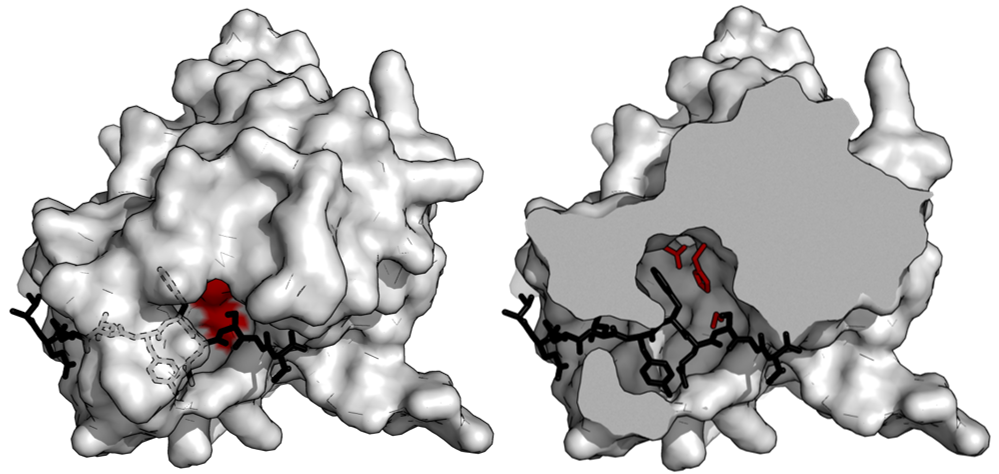
| + | [[File:TEV2.png|none|400px|thumb|wikipedia: ''Surface model of TEV bound to uncleaved substrate (black), also showing the catalytic triad (red). The substrate binds inside an active site tunnel (left). A cutaway (right) shows the complementary shape of the binding tunnel to the substrate. (PDB: 1lvb)'']] | ||
| + | |||
| + | [[File:TEV1.png|none|400px|thumb|Waugh DS wrote: ''TEV protease sequence specificity is far more stringent than that of factor Xa, thrombin, or enterokinase. It is a very useful reagent for cleaving fusion proteins. TEV protease recognizes a linear epitope of the general form E-Xaa-Xaa-Y-Xaa-Q-(G/S), with cleavage occurring between Q and G or Q and S.'']] | ||
| + | Please refer to [[Media:Tev-faq-7pages.pdf]] for more details on TEV protease. | ||
| + | |||
| + | Note: [[Part:BBa_K2549013]], [[Part:BBa_K2549014]] and [[Part:BBa_K2549015]] have the same Applications section. | ||
| Line 17: | Line 24: | ||
<partinfo>BBa_K2549013 parameters</partinfo> | <partinfo>BBa_K2549013 parameters</partinfo> | ||
--> | --> | ||
| − | |||
===References=== | ===References=== | ||
Revision as of 13:47, 7 October 2018
TEV protease
This part is a mutant TEV (tobacco etch virus) protease. The residue 219 of the wild-type TEV protease is mutated from serine to valine (S219V) to remove autoinactivation and perform a more efficient cleavage[1]. TEV protease is one of the best-characterized enzymes of the viral proteases which have more stringent sequence specificity. We used it to build our IMPLY logic gates. It can also be utilized by other iGEM teams to create their own engineered fusion proteins cleavage system.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 316
Applications
The main use of TEV protease[2] is for removing affinity tags from purified recombinant fusion proteins. The reason for that is due to the high sequence specificity. This specificity allows for the controlled cleavage of proteins when the preference sequence is inserted into flexible loops. It also makes it relatively non-toxic in vivo as the recognized sequence scarcely occurs in proteins.
Please refer to Media:Tev-faq-7pages.pdf for more details on TEV protease.
Note: Part:BBa_K2549013, Part:BBa_K2549014 and Part:BBa_K2549015 have the same Applications section.
References
- ↑ Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Kapust RB, Tözsér J, Fox JD, ..., Copeland TD, Waugh DS. Protein Eng, 2001 Dec;14(12):993-1000 PMID: 11809930
- ↑ https://en.wikipedia.org/wiki/TEV_protease