Difference between revisions of "Part:BBa K2812000"
| Line 3: | Line 3: | ||
<partinfo>BBa_K2812000 short</partinfo> | <partinfo>BBa_K2812000 short</partinfo> | ||
| − | Carbohydrate binding domain from the Marinomonas primoryensis ice-binding protein ( | + | Carbohydrate binding domain from the ''Marinomonas primoryensis'' ice-binding protein (''Mp''IBP). It requires millimolar concentrations of calcium ions to properly fold into a globular β-fold. A coordinated calcium ion is used to bind sugar moieties, such as glucose. Unfolding of the domain can be induced by the addition of EDTA, which chelates the calcium ions and prevents binding of sugar moieties. It does not require the presence of other domains to fold and therefore it can be used as a modular protein domain to bind sugar moieties. TU Eindhoven 2018 used the carbohydrate binding domain from ''Mp''IBP to tether ''E. coli'' bacteria to dextran, a glucose-based polymer. |
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | The Marinomonas primoryensis ice-binding protein | + | The ''Marinomonas primoryensis'' ice-binding protein (''Mp''IBPis a 1.5-MDa adhesin used by the Antarctic bacterium ''M. primoryensis'' to bind to ice and diatoms to position itself on top of the water column to access nutrients and oxygen. The carbohydrate binding domain from this protein is used to bind extracellular polysaccharides to form microcolonies and to tether ''M. primoryensis'' to other photosynthetic microorganisms. |
==Proof of Functionality== | ==Proof of Functionality== | ||
====Experimental Setup==== | ====Experimental Setup==== | ||
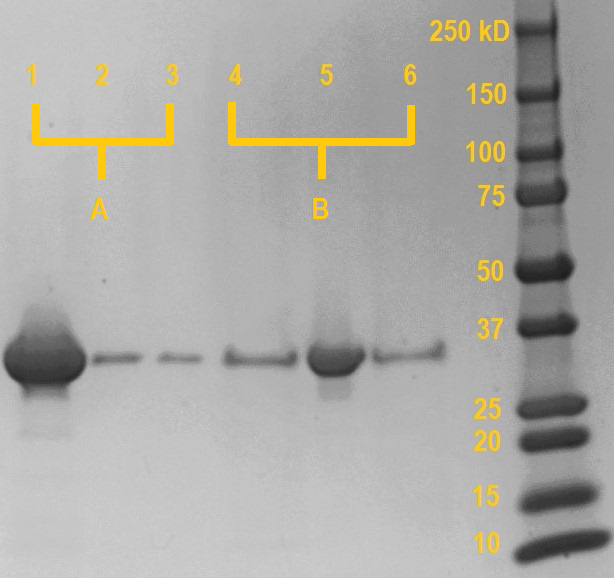
| − | This experiment was performed to verify the calcium-dependent carbohydrate binding capabilities of the | + | This experiment was performed to verify the calcium-dependent carbohydrate binding capabilities of the ''Mp''IBP carbohydrate binding domain. Equimolar amounts purified ''Mp''IBP carbohydrate binding domain were incubated with Superdex dextran beads in the presence of 5mM CaCl2 (sample A) or 10mM EDTA (Sample B). Dextran is a glucose polymer, to which our ''Mp''IBP carbohydrate binding domain should bind in the presence of millimolar concentrations of calcium. After incubation, each sample was centrifuged shortly at 8,000 rpm to pull the dextran beads (and any dextran-bound proteins) down from the mixture and the supernatant was isolated. Next, the beads were resuspended in buffer without CaCl2 (sample A) or with EDTA (sample B), allowed to incubate. After incubation, the samples were centrifuged and the supernatant was isolated. |
SDS samples were prepared from the dextran beads after the final centrifugation (lane 1 & 4), supernatant from the first incubation (lane 2 & 5) and supernatant from the resuspension (lane 3 & 6). The samples were boiled at 95 degrees Celcius for 15 minutes and the SDS gel was run at 120V for one hour. Anything bound to the dextran beads will be released into the solution during the boiling. | SDS samples were prepared from the dextran beads after the final centrifugation (lane 1 & 4), supernatant from the first incubation (lane 2 & 5) and supernatant from the resuspension (lane 3 & 6). The samples were boiled at 95 degrees Celcius for 15 minutes and the SDS gel was run at 120V for one hour. Anything bound to the dextran beads will be released into the solution during the boiling. | ||
| Line 16: | Line 16: | ||
====Results==== | ====Results==== | ||
[[File:BBa_K2812000_CBD_Assay_detailed.png|300px|right|SDS PAGE gel from the Carbohydrate Binding Assay|thumb]] | [[File:BBa_K2812000_CBD_Assay_detailed.png|300px|right|SDS PAGE gel from the Carbohydrate Binding Assay|thumb]] | ||
| − | As can be seen on the SDS gel on the right, if the | + | As can be seen on the SDS gel on the right, if the ''Mp''IBP carbohydrate binding domain is incubated with dextran in the presence of millimolar concentrations of calcium, only a very small amount of protein remains unbound in the supernatant (lane 2; probably due to saturation of the dextran beads). However, if the protein domain is incubated in the presence of EDTA, which chelates the calcium, almost all protein remains in unbound in the supernatant (lane 5). |
In the subsequent washing step (lane 3 & 6), aspecifically bound protein is washed away from the beads using buffer devoid of calcium. For both sample A and B, the amount of aspecifically bound protein released during the washing is very minor and comparable. This shows that most proteins stay bound in the case of sample A, while in sample B most protein has already been removed in the previous step. | In the subsequent washing step (lane 3 & 6), aspecifically bound protein is washed away from the beads using buffer devoid of calcium. For both sample A and B, the amount of aspecifically bound protein released during the washing is very minor and comparable. This shows that most proteins stay bound in the case of sample A, while in sample B most protein has already been removed in the previous step. | ||
| Line 23: | Line 23: | ||
====Conclusion==== | ====Conclusion==== | ||
| − | The observations above show that the binding of the | + | The observations above show that the binding of the ''Mp''IBP carbohydrate binding domain is calcium dependent and that negligible binding occurs in the absence of calcium. Once bound in the presence of calcium, there is no need for additional calcium to remain present in the solution as the protein-carbohydrate complex holds on to the calcium ion tightly. EDTA can be used to chelate the calcium, inducing unfolding and prevent binding of the protein. |
| − | + | ||
| − | + | ||
| + | The isolated carbohydrate binding domain of the 1.5 MDa ''Mp''IBP has been proven to retain its functionality in the absence of the rest of the adhesin. | ||
<!-- --> | <!-- --> | ||
Revision as of 16:36, 23 September 2018
Carbohydrate-binding domain from MpIBP
Carbohydrate binding domain from the Marinomonas primoryensis ice-binding protein (MpIBP). It requires millimolar concentrations of calcium ions to properly fold into a globular β-fold. A coordinated calcium ion is used to bind sugar moieties, such as glucose. Unfolding of the domain can be induced by the addition of EDTA, which chelates the calcium ions and prevents binding of sugar moieties. It does not require the presence of other domains to fold and therefore it can be used as a modular protein domain to bind sugar moieties. TU Eindhoven 2018 used the carbohydrate binding domain from MpIBP to tether E. coli bacteria to dextran, a glucose-based polymer.
Usage and Biology
The Marinomonas primoryensis ice-binding protein (MpIBPis a 1.5-MDa adhesin used by the Antarctic bacterium M. primoryensis to bind to ice and diatoms to position itself on top of the water column to access nutrients and oxygen. The carbohydrate binding domain from this protein is used to bind extracellular polysaccharides to form microcolonies and to tether M. primoryensis to other photosynthetic microorganisms.
Proof of Functionality
Experimental Setup
This experiment was performed to verify the calcium-dependent carbohydrate binding capabilities of the MpIBP carbohydrate binding domain. Equimolar amounts purified MpIBP carbohydrate binding domain were incubated with Superdex dextran beads in the presence of 5mM CaCl2 (sample A) or 10mM EDTA (Sample B). Dextran is a glucose polymer, to which our MpIBP carbohydrate binding domain should bind in the presence of millimolar concentrations of calcium. After incubation, each sample was centrifuged shortly at 8,000 rpm to pull the dextran beads (and any dextran-bound proteins) down from the mixture and the supernatant was isolated. Next, the beads were resuspended in buffer without CaCl2 (sample A) or with EDTA (sample B), allowed to incubate. After incubation, the samples were centrifuged and the supernatant was isolated.
SDS samples were prepared from the dextran beads after the final centrifugation (lane 1 & 4), supernatant from the first incubation (lane 2 & 5) and supernatant from the resuspension (lane 3 & 6). The samples were boiled at 95 degrees Celcius for 15 minutes and the SDS gel was run at 120V for one hour. Anything bound to the dextran beads will be released into the solution during the boiling.
Results
As can be seen on the SDS gel on the right, if the MpIBP carbohydrate binding domain is incubated with dextran in the presence of millimolar concentrations of calcium, only a very small amount of protein remains unbound in the supernatant (lane 2; probably due to saturation of the dextran beads). However, if the protein domain is incubated in the presence of EDTA, which chelates the calcium, almost all protein remains in unbound in the supernatant (lane 5).
In the subsequent washing step (lane 3 & 6), aspecifically bound protein is washed away from the beads using buffer devoid of calcium. For both sample A and B, the amount of aspecifically bound protein released during the washing is very minor and comparable. This shows that most proteins stay bound in the case of sample A, while in sample B most protein has already been removed in the previous step.
Boiling the dextran beads releases any protein still bound after washing. In the case of sample A (initially incubated in the presence of calcium), a very large amount of protein has remained bound during the washing and is released from the beads (lane 1). Conversely, the beads incubated with the protein in the presence of EDTA release negligible amounts of protein compared to sample A, as almost all protein has already been washed away.
Conclusion
The observations above show that the binding of the MpIBP carbohydrate binding domain is calcium dependent and that negligible binding occurs in the absence of calcium. Once bound in the presence of calcium, there is no need for additional calcium to remain present in the solution as the protein-carbohydrate complex holds on to the calcium ion tightly. EDTA can be used to chelate the calcium, inducing unfolding and prevent binding of the protein.
The isolated carbohydrate binding domain of the 1.5 MDa MpIBP has been proven to retain its functionality in the absence of the rest of the adhesin.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]