Difference between revisions of "Part:BBa K1399011"
Djcamenares (Talk | contribs) m |
|||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1399011 short</partinfo> | <partinfo>BBa_K1399011 short</partinfo> | ||
Lactose/IPTG inducible promoter with RBS, RFP reporter tagged with LVA-ssrA degradation tag and terminator. | Lactose/IPTG inducible promoter with RBS, RFP reporter tagged with LVA-ssrA degradation tag and terminator. | ||
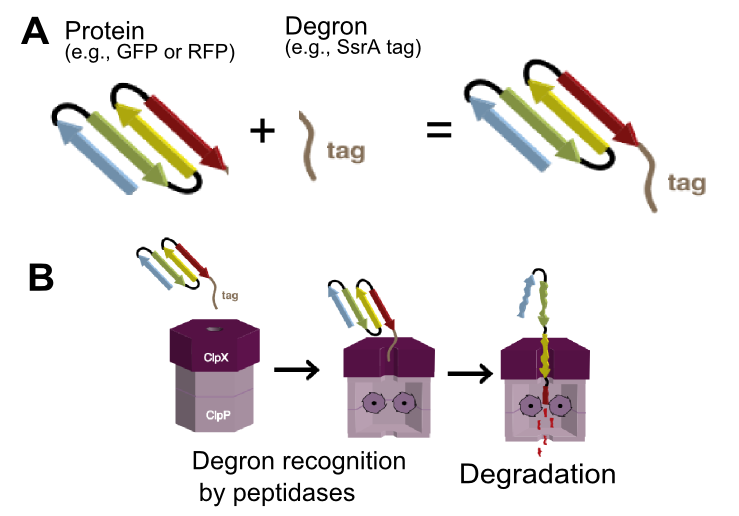
| − | The tagged RFP is actively degraded within cell, thus provides better temporal resolution of red fluorescence and promoter activity. | + | The tagged RFP is actively degraded within cell (Figure 1), thus provides better temporal resolution of red fluorescence and promoter activity. |
| − | This part can be considered as improved version of BBa_J04451 as it does not contain StuI restriction site coding for additional 'RP' residues between RFP and LVA tag. | + | This part can be considered as improved version of [[Part:BBa_J04451]] as it does not contain StuI restriction site coding for additional 'RP' residues between RFP and LVA tag. |
| + | [[File:EDiGEM14-SsrA degron mediated degradation.png|400px|thumb|center|'''Figure 1 SsrA degron mediated protein degradation.''' ('''A''') Any version of SsrA tags can be attached to any protein of interest (e.g. RFP or GFP). ('''B''') The tag is recognized by ClpX unfoldase forming a complex with ClpP protease and the tagged protein gets degraded.]] | ||
| − | <!-- Add more about the biology of this part here | + | <!-- Add more about the biology of this part here --> |
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | |||
| + | [http://2017.igem.org/Team:Kingsborough_NY Team Kingsborough NY 2017] contributed to the characterization of this part by showing decreased fluorescence when expressed either in a higher salt media - such as LB with 3% sodium chloride - or E. coli that lacks tmRNA, the principal component of the cell's ribosome rescue system. Cells without tmRNA will have less native ssrA substrates, and this may lead to increased degradation of ssrA tagged RFP. For more information and data, see the experience page or [http://2017.igem.org/Team:Kingsborough_NY/RFP visit our Wiki]<br> | ||
| + | (<small>--[[User:djcamenares|djcamenares]] 17:56, 27 October 2017 (UTC) </small>) | ||
<!-- --> | <!-- --> | ||
Latest revision as of 19:55, 19 November 2017
Plac-RFP(LVA)
Lactose/IPTG inducible promoter with RBS, RFP reporter tagged with LVA-ssrA degradation tag and terminator. The tagged RFP is actively degraded within cell (Figure 1), thus provides better temporal resolution of red fluorescence and promoter activity. This part can be considered as improved version of Part:BBa_J04451 as it does not contain StuI restriction site coding for additional 'RP' residues between RFP and LVA tag.
Usage and Biology
[http://2017.igem.org/Team:Kingsborough_NY Team Kingsborough NY 2017] contributed to the characterization of this part by showing decreased fluorescence when expressed either in a higher salt media - such as LB with 3% sodium chloride - or E. coli that lacks tmRNA, the principal component of the cell's ribosome rescue system. Cells without tmRNA will have less native ssrA substrates, and this may lead to increased degradation of ssrA tagged RFP. For more information and data, see the experience page or [http://2017.igem.org/Team:Kingsborough_NY/RFP visit our Wiki]
(--djcamenares 17:56, 27 October 2017 (UTC) )
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 781
Illegal AgeI site found at 893 - 1000COMPATIBLE WITH RFC[1000]