Difference between revisions of "Part:BBa K1399012:Experience"
Djcamenares (Talk | contribs) |
|||
| (One intermediate revision by one other user not shown) | |||
| Line 2: | Line 2: | ||
This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | ||
how you used this part and how it worked out. | how you used this part and how it worked out. | ||
| + | |||
| + | == '''Team Kingsborough NY 2017''' == | ||
| + | |||
| + | Proteins with a SsrA tag, or some variant of this tag, are degraded by cellular proteases such as ClpXP. However, the rate of degradation for a tagged reporter (such as RFP-ssrA, K1399012, or similar variants) will depend upon the level of endogenous SsrA-tagging that occurs - this tag is added in response to ribosome rescue. Therefore, we hypothesize that abolishing the tmRNA ribosome rescue system which adds the SsrA tag to proteins will destabilize a genetically tagged reporter, since it will now be the only target present for cellular proteases (by corollary, increased SsrA-tagging in response to some stress should stabilize a SsrA tagged reporter). | ||
| + | |||
| + | To test this, we transformed constructs K1399010 through K1399013, or J04450 (a control, RFP with no tag) into either wild-type (WT) or tmRNA deficient cells (<i>ssrA::Kan-R</i>). Decreased RFP levels were observed in strains without tmRNA, which is consistent with our hypothesis. However, there are alternative explanations, as deletion of <i>ssrA</i> does effect the dynamics and steady state levels of expression from a LacI-responsive promoter, as we demonstrated on the [https://parts.igem.org/Part:BBa_J04450:Experience#Team_Kingsborough_NY_2017 experience page for BBa_J04550]. | ||
| + | |||
| + | [[File:T--Kingsborough_NY--RFP_ssrA_fig1.png|800px]] | ||
| + | |||
| + | <i>Fig 1: E. coli, either wild-type (WT) or trans-translation deficient (ssrA::Kan-R) with RFP variants streaked onto LB Miller + Chloramphenicol plates, and in some cases supplemented with 1mM IPTG, and grown for 1 day at 37C. This image is a representative of multiple experiments.</i> | ||
| + | |||
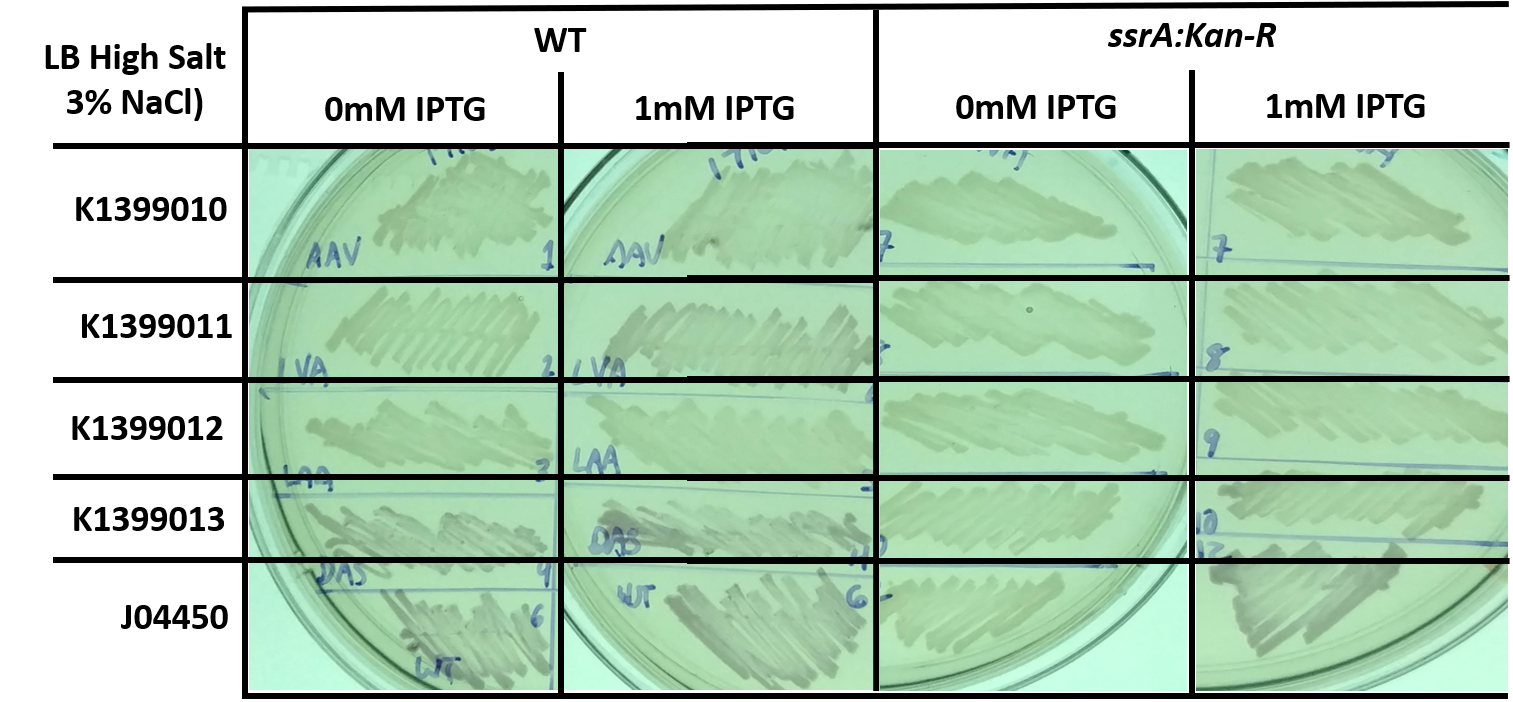
| + | In connection with another experiment, we also tested the same constructs in the same backgrounds but grown on LB with a higher salt concentration, similar to that which is found in seawater. However, the overall decrease in expression makes it difficult to compare the different constructs and genetic background. | ||
| + | |||
| + | [[File:T--Kingsborough_NY--RFP_ssrA_fig3.png|800px]] | ||
| + | |||
| + | <i>Fig 2: E. coli, either wild-type (WT) or trans-translation deficient (ssrA::Kan-R) with RFP variants streaked onto high salt LB (3% NaCl) + Chloramphenicol plates, and in some cases supplemented with 1mM IPTG, and grown for 1 day at 37C. This image is a representative of multiple experiments.</i> | ||
===Applications of BBa_K1399012=== | ===Applications of BBa_K1399012=== | ||
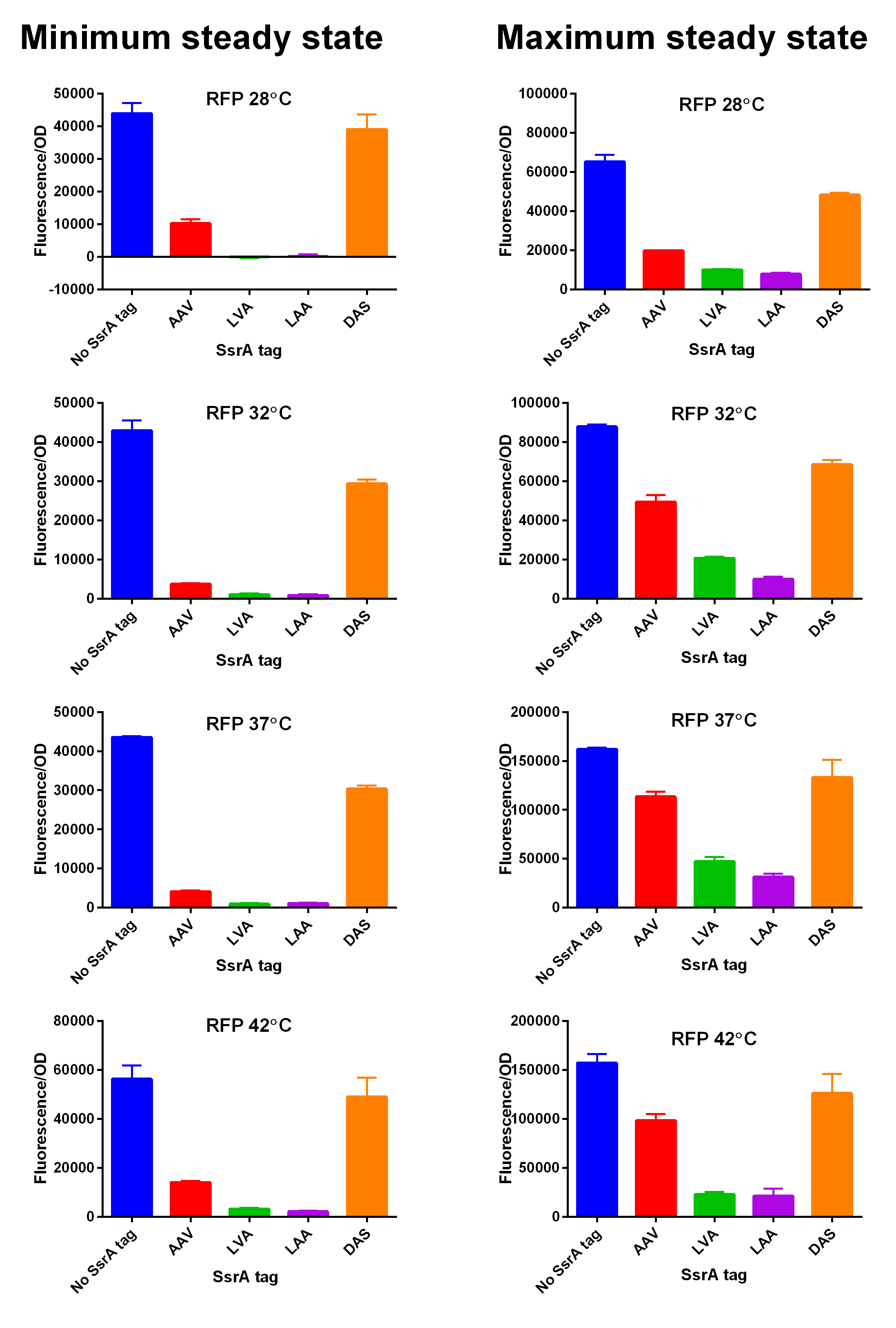
| + | We characterized and compared the instability of this SsrA-LAA tagged RFP to others containing different variants of SsrA tags. We determined the steady state fluorescence/OD ratio of E. coli JM109 at uninduced (minimum steady state) and induced (maximum steady state) state. To see how the temperature of environment can affect protein degradation efficiency we repeated experiments in several temperatures (28⁰C, 32⁰C, 37⁰C and 42⁰C). Our data are summarised below. (For more information on SsrA degradation tag system, our experimental assays and results please visit our wiki page [http://2014.igem.org/Team:Edinburgh].) | ||
| + | [[File:Tagged RFP min and max values at different temperatures.jpg|600px|thumb|center|'''SsrA degron tagged RFP minimum (left) and maximum (right) fluorescence per OD at four different temperatures (28⁰C, 32⁰C, 37⁰C and 42⁰C).''']] [[File:EDiGEM14 RFP legend.png|500px|thumb|center|'''Summary of BioBricks to which our characterization data is applicable.''']] | ||
===User Reviews=== | ===User Reviews=== | ||
Latest revision as of 19:54, 19 November 2017
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Team Kingsborough NY 2017
Proteins with a SsrA tag, or some variant of this tag, are degraded by cellular proteases such as ClpXP. However, the rate of degradation for a tagged reporter (such as RFP-ssrA, K1399012, or similar variants) will depend upon the level of endogenous SsrA-tagging that occurs - this tag is added in response to ribosome rescue. Therefore, we hypothesize that abolishing the tmRNA ribosome rescue system which adds the SsrA tag to proteins will destabilize a genetically tagged reporter, since it will now be the only target present for cellular proteases (by corollary, increased SsrA-tagging in response to some stress should stabilize a SsrA tagged reporter).
To test this, we transformed constructs K1399010 through K1399013, or J04450 (a control, RFP with no tag) into either wild-type (WT) or tmRNA deficient cells (ssrA::Kan-R). Decreased RFP levels were observed in strains without tmRNA, which is consistent with our hypothesis. However, there are alternative explanations, as deletion of ssrA does effect the dynamics and steady state levels of expression from a LacI-responsive promoter, as we demonstrated on the experience page for BBa_J04550.
Fig 1: E. coli, either wild-type (WT) or trans-translation deficient (ssrA::Kan-R) with RFP variants streaked onto LB Miller + Chloramphenicol plates, and in some cases supplemented with 1mM IPTG, and grown for 1 day at 37C. This image is a representative of multiple experiments.
In connection with another experiment, we also tested the same constructs in the same backgrounds but grown on LB with a higher salt concentration, similar to that which is found in seawater. However, the overall decrease in expression makes it difficult to compare the different constructs and genetic background.
Fig 2: E. coli, either wild-type (WT) or trans-translation deficient (ssrA::Kan-R) with RFP variants streaked onto high salt LB (3% NaCl) + Chloramphenicol plates, and in some cases supplemented with 1mM IPTG, and grown for 1 day at 37C. This image is a representative of multiple experiments.
Applications of BBa_K1399012
We characterized and compared the instability of this SsrA-LAA tagged RFP to others containing different variants of SsrA tags. We determined the steady state fluorescence/OD ratio of E. coli JM109 at uninduced (minimum steady state) and induced (maximum steady state) state. To see how the temperature of environment can affect protein degradation efficiency we repeated experiments in several temperatures (28⁰C, 32⁰C, 37⁰C and 42⁰C). Our data are summarised below. (For more information on SsrA degradation tag system, our experimental assays and results please visit our wiki page [http://2014.igem.org/Team:Edinburgh].)
User Reviews
UNIQ37f47361ca08ae75-partinfo-00000000-QINU UNIQ37f47361ca08ae75-partinfo-00000001-QINU