Difference between revisions of "Part:BBa K2457004"
| (18 intermediate revisions by 5 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2457004 short</partinfo> | <partinfo>BBa_K2457004 short</partinfo> | ||
| − | + | <p>This BioBrick is a key building block of the CRISPeasy-in (CRIN) framework, developed in 2017 by the Amazonas_Brazil team, aiming to implement bioengineering principles into CRISPR based methods. Our proposal was to rationally design parts to be interchangeable and standardized, providing a BioBrick toolbox to attend multiple SynBio purposes. For that matter, this BioBrick was designed to provide a novel Donor DNA template system to integrate BioBricks into bacterial genome with RFC10 compatibility. It's composed of a GFP gene, flanked by NheI restriction sites. It's versatility and novelty relies on the possibility to switch by one-step cloning the GFP for any BioBrick of interest. It's the first available on Registry BioBrick for a Donor DNA template system.</p> | |
[[File: BBa K2457004 circuit.png]] | [[File: BBa K2457004 circuit.png]] | ||
| Line 11: | Line 11: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | Composed by the JK26 promoter, the coding sequence | + | <p>Composed by: the JK26 promoter, the coding sequence for the Green Fluorescent Protein (GFP) and the ilvBN terminator.</p> |
| − | <p> | + | |
| − | === | + | <p>It has 1810bp and was designed to contain two homologous arms with 300bp from the LacZ gene located on the Operon Lac. According to Zhao et al (2016), the use of recombination arms with 300bp increases the efficiency in 100%. On the other hand, if you have a 150bp arm, the efficiency decreases to 63%.</p> |
| + | When the homologous arms are not big enough, that's what happens: | ||
| + | The efficiency decays abruptly from 100% to 3%~7% range; there are two cell repair pathways after the cleavage by Cas9: HDR and NHEJ. NHEJ is a system that repairs without a recombinase action and simply binds the extremities. HDR is the repair which uses the recombinases action; and If the homologous arms are too short, it is easier to NHEJ happens. | ||
| + | |||
| + | <p> Between the homologous arms, lies the GFP gene, flanked by NheI sites, allowing an RFC10 versatile one-step compatibility, the insertion of any other gene in GFP site.</p> | ||
| + | |||
| + | <p>It has this structure especially so the DSB repair will be guided to the HDR (Homology Direct Repair), where homology is recognized by the RecA enzyme, which will break through the BioBrick to restore the cleaved sequence in the genome, having the aimed gene integrated.</p> | ||
| + | |||
| + | ===Assembly=== | ||
| + | The sequence was ordered as gBlocks from Integrated DNA Technologies and it was assembled by restriction enzyme site. | ||
| + | |||
| + | [[File:Donor_Dig.jpg]] | ||
| + | |||
| + | Figure 2: Eletrophoretic profile of restriction digestion of BBa_K2457004. | ||
===Characterization=== | ===Characterization=== | ||
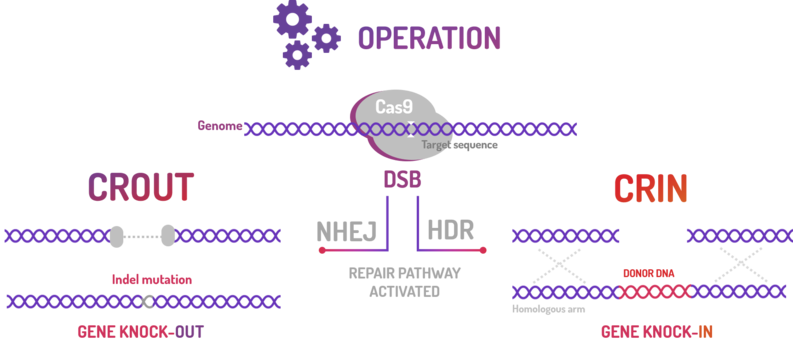
| + | <p>BBa_K2457004 was used by Amazonas_Brazil team in one method based on standard BioBricks modules for bacterial genome engineering mediated by CRISPR-Cas9 machinery: the CRISPeasys. The approaches constructed were 1) for non-homologous end joining (NHEJ) repair pathway, the CRISPeasy-out method (or crOUT), referring to knock-out and the 2) for homologous directed repair (HDR), the CRISPeasy-in method (or crIN), referring to knock-in. <b>This part was used especially for the second approach (CRIN).</b> </p> | ||
| + | [[File:CROUT CRIN 1.png]] | ||
| + | <p>Figure 3: CRISPeasy subdivisions.</p> | ||
| + | <p><b>2. CRIN (CRISPeasy-in method): Designed for knock-in strategy for bacterial genome editing based on CRISPR/Cas9</b></p> | ||
| + | |||
| + | <p>As the main key to this method, there is an AND logic gate composed of Cas9 coupled with RecA, which operates the Donor DNA insertion into E. coli genome through Direct Homology Repair (HDR). In this way, the arabinose AND IPTG (the inputs) activate the gene expression program encoding Cas9 and RecA. Then, there is the execution of their hardware's (physical parts, e.g. proteins) operation. As an output, Cas9 AND RecA activate the DNA insertion into the genome. | ||
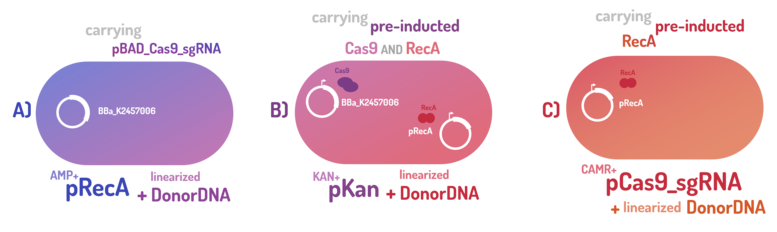
| + | In order to standardize a reliable datasheet of genome engineering efficiency, we tested a range of electrocompetent <i>E. coli</i> K12 MG1655 cells with different circuits: test A) carrying a non-pre-inducted Cas9 device + sgRNA (BBa_K2457006) plasmid; B) carrying a pre-inducted Cas9 AND RecA; C) carrying a pre-inducted RecA. The donor DNA was electroporated in all the tests. </p> | ||
| + | |||
| + | [[File:CRIN 2 METHODOLOGY.png]] | ||
| + | |||
| + | Figure 4: CRISPeasy-in methodology. | ||
| + | <p><b>A) carrying a non-pre-inducted Cas9 device + sgRNA;</b></p> | ||
| + | <p>We transformed through electroporation <i>E. coli</i> K12 MG1655 carrying a non-pre-inducted pBAD_Cas9_sgRNA plasmid CamR+ (BBa_K2457006) with pRecA (AmpR+) and a linearized Donor DNA (BBa_K2457004). We screened over 94 transformants colonies, inoculating in 200µL of LB + 25µg/mL chloramphenicol + 100µg/mL ampicillin + 0.2% L-Arabinose + 1mM IPTG to activate the expression of RecA AND Cas9. We measured the initial OD (600nm) and initial fluorescence (exc. 340 nm em. 530 nm) on 96-wells plates applying the Hidex Chameleon V spectrofluorimeter. The process was repeated for each hour over 10 hours while the 96-wells plate kept growing on a shaker at 37ºC.</p> | ||
| + | |||
| + | <p><b>B) carrying a pre-inducted Cas9 + sgRNA AND RecA;</b></p> | ||
| + | |||
| + | <p>We transformed through electroporation <i>E. coli</i> K12 MG1655 carrying a pre-inducted pBAD_Cas9_sgRNA plasmid CamR+ (BBa_K2457006) AND pRecA plasmid AmpR+ with a linearized Donor DNA (BBa_K2457004) + pKana. We screened over 94 transformants colonies, inoculating in 200µL of LB + 50µg/mL kanamycin + 25µg/mL chloramphenicol + 100µg/mL ampicillin + 0.2% L-Arabinose + 1mM IPTG to activate the expression of RecA AND Cas9. We measured the initial OD (600nm) and initial fluorescence (exc. 340 nm em. 530 nm) on 96-wells plates applying the Hidex Chameleon V spectrofluorimeter. The process was repeated for each hour over 10 hours while the 96-wells plate kept growing on a shaker at 37ºC.</p> | ||
| + | |||
| + | <p><b>C) carrying a pre-inducted RecA.</b></p> | ||
| + | <p>We transformed through electroporation <i>E. coli</i> K12 MG1655 carrying a pRecA (AmpR+) with pBAD_Cas9_sgRNA plasmid CamR+ (BBa_K2457006) and a linearized Donor DNA (BBa_K2457004). We screened over 80 transformants colonies, inoculating in 200µL of LB + 25µg/mL chloramphenicol + 100µg/mL ampicillin + 0.2% L-Arabinose + 1mM IPTG to activate the expression of RecA AND Cas9. We measured the initial OD (600nm) and initial fluorescence (exc. 340 nm em. 530 nm) on 96-wells plates applying the Hidex Chameleon V spectrofluorimeter. The process was repeated for each hour over 10 hours while the 96-wells plate kept growing on a shaker at 37ºC.</p> | ||
| + | |||
| + | |||
| + | [[File:CRIN 1.png]] | ||
| + | |||
| + | [[File:CRIN 2.png]] | ||
| + | |||
| + | [[File:CRIN 3.png]] | ||
| + | |||
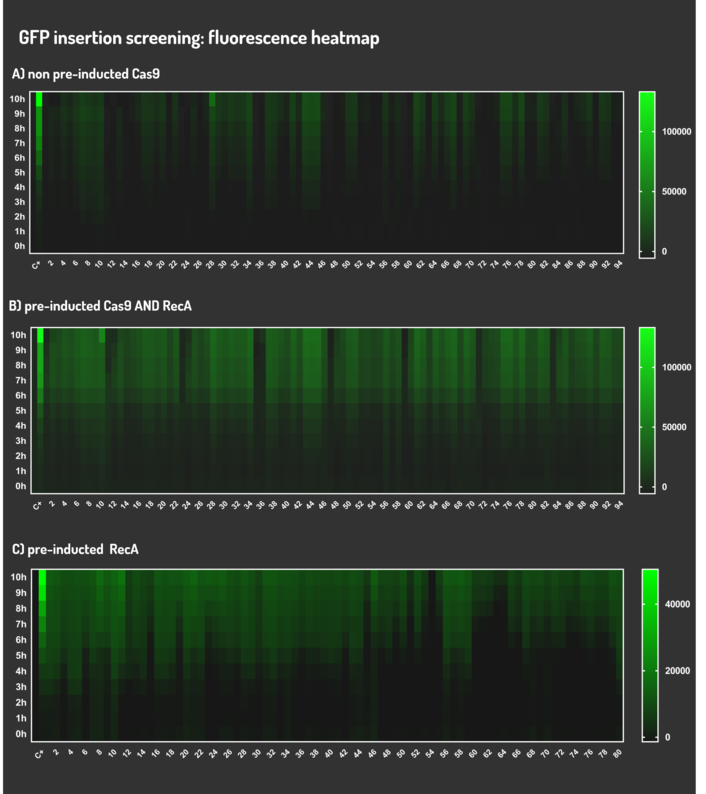
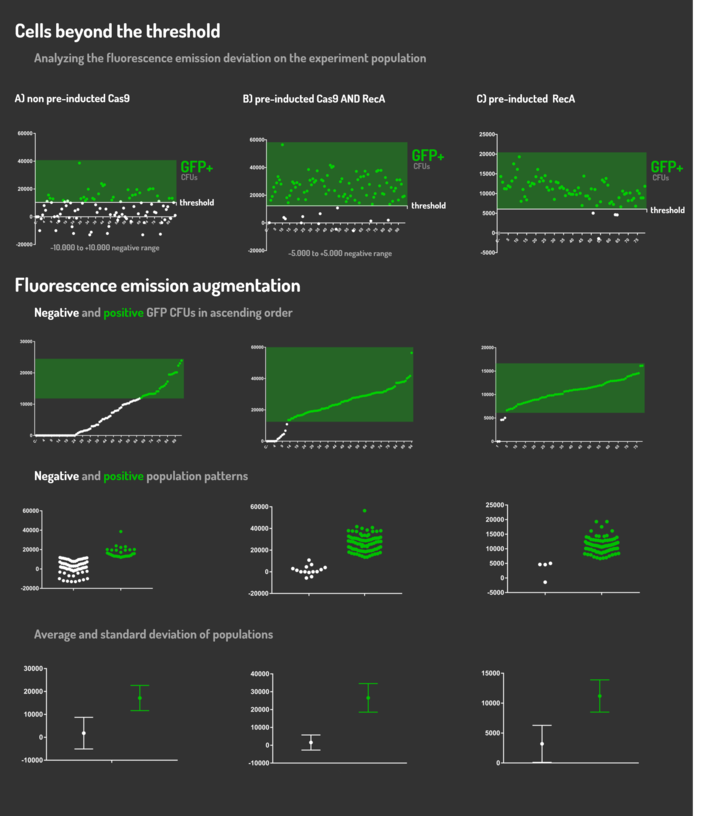
| + | <p>Figure 5: CRISPeasy-in results.</p> | ||
| + | <p>Leading from the achieved results, it was noted that has certain variations between the methodologies performances: with non-induced Cas9 (30% recombinants), with pre-induced Cas9 and RecA (90% recombinants) and with exclusively pre-induced RecA (95% recombinants). </p> | ||
| + | <p>The non-induced Cas9 method possibly do not have the best performance (30%) due to the absence of Cas9 protein when the Donor DNA is inserted into genome, causing a delay in the target sequence cleavage by Cas9, and, therefore, also the delay in homologous recombination, then the major linear Donor DNA is degraded in the bacterial cytosol.</p> | ||
| + | <p>The pre-induced Cas9 and RecA presented a very good result and similar to the most effective approach. When Donor DNA is inserted into the genome, is already happening the cleavage by Cas9 and RecA is investigating a homologous sequence to the broken strand, then Donor DNA is quickly incorporated, having less DNA degradation. </p> | ||
| + | <p>We found out that the most efficient method was the one which had only RecA pre-induced, since Cas9 presence affects the cells growth, therefore, when Cas9 and Donor DNA are added simultaneously, it will have the normal cell growth, beyond the further homologous recombination. </p> | ||
| + | <p>The CRISPeasy framework development can be a boost for an easily AND reliable way to use bacterial genome editing based on CRISPR/Cas9. We encourage teams to dive into CRISPR's standardization with us and expect to be able to see in the next years more and more projects with technique's improvement!</p> | ||
===Sequencing=== | ===Sequencing=== | ||
[[File:BBa K2457004 electropherogram.png]] | [[File:BBa K2457004 electropherogram.png]] | ||
| − | Figure | + | Figure 6:Sequencing electropherogram from BBa_K2457004 |
[[File:BBa K2457004 aligmnent.png]] | [[File:BBa K2457004 aligmnent.png]] | ||
| − | Figure | + | Figure 7:Alignment of the designed sequence and our final construction from BBa_K2457004. |
<span class='h3bb'>Sequence and Features</span></p> | <span class='h3bb'>Sequence and Features</span></p> | ||
Latest revision as of 03:27, 2 November 2017
Donor DNA for HDR into LacZ
This BioBrick is a key building block of the CRISPeasy-in (CRIN) framework, developed in 2017 by the Amazonas_Brazil team, aiming to implement bioengineering principles into CRISPR based methods. Our proposal was to rationally design parts to be interchangeable and standardized, providing a BioBrick toolbox to attend multiple SynBio purposes. For that matter, this BioBrick was designed to provide a novel Donor DNA template system to integrate BioBricks into bacterial genome with RFC10 compatibility. It's composed of a GFP gene, flanked by NheI restriction sites. It's versatility and novelty relies on the possibility to switch by one-step cloning the GFP for any BioBrick of interest. It's the first available on Registry BioBrick for a Donor DNA template system.
Figure 1: BBa K2457004 circuit
Usage and Biology
Composed by: the JK26 promoter, the coding sequence for the Green Fluorescent Protein (GFP) and the ilvBN terminator.
It has 1810bp and was designed to contain two homologous arms with 300bp from the LacZ gene located on the Operon Lac. According to Zhao et al (2016), the use of recombination arms with 300bp increases the efficiency in 100%. On the other hand, if you have a 150bp arm, the efficiency decreases to 63%.
When the homologous arms are not big enough, that's what happens: The efficiency decays abruptly from 100% to 3%~7% range; there are two cell repair pathways after the cleavage by Cas9: HDR and NHEJ. NHEJ is a system that repairs without a recombinase action and simply binds the extremities. HDR is the repair which uses the recombinases action; and If the homologous arms are too short, it is easier to NHEJ happens.
Between the homologous arms, lies the GFP gene, flanked by NheI sites, allowing an RFC10 versatile one-step compatibility, the insertion of any other gene in GFP site.
It has this structure especially so the DSB repair will be guided to the HDR (Homology Direct Repair), where homology is recognized by the RecA enzyme, which will break through the BioBrick to restore the cleaved sequence in the genome, having the aimed gene integrated.
Assembly
The sequence was ordered as gBlocks from Integrated DNA Technologies and it was assembled by restriction enzyme site.
Figure 2: Eletrophoretic profile of restriction digestion of BBa_K2457004.
Characterization
BBa_K2457004 was used by Amazonas_Brazil team in one method based on standard BioBricks modules for bacterial genome engineering mediated by CRISPR-Cas9 machinery: the CRISPeasys. The approaches constructed were 1) for non-homologous end joining (NHEJ) repair pathway, the CRISPeasy-out method (or crOUT), referring to knock-out and the 2) for homologous directed repair (HDR), the CRISPeasy-in method (or crIN), referring to knock-in. This part was used especially for the second approach (CRIN).
Figure 3: CRISPeasy subdivisions.
2. CRIN (CRISPeasy-in method): Designed for knock-in strategy for bacterial genome editing based on CRISPR/Cas9
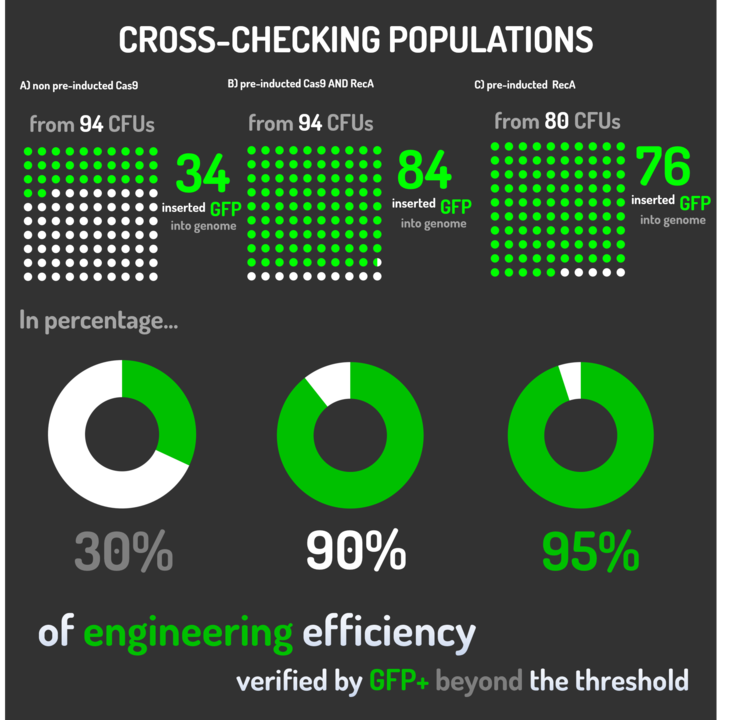
As the main key to this method, there is an AND logic gate composed of Cas9 coupled with RecA, which operates the Donor DNA insertion into E. coli genome through Direct Homology Repair (HDR). In this way, the arabinose AND IPTG (the inputs) activate the gene expression program encoding Cas9 and RecA. Then, there is the execution of their hardware's (physical parts, e.g. proteins) operation. As an output, Cas9 AND RecA activate the DNA insertion into the genome. In order to standardize a reliable datasheet of genome engineering efficiency, we tested a range of electrocompetent E. coli K12 MG1655 cells with different circuits: test A) carrying a non-pre-inducted Cas9 device + sgRNA (BBa_K2457006) plasmid; B) carrying a pre-inducted Cas9 AND RecA; C) carrying a pre-inducted RecA. The donor DNA was electroporated in all the tests.
Figure 4: CRISPeasy-in methodology.
A) carrying a non-pre-inducted Cas9 device + sgRNA;
We transformed through electroporation E. coli K12 MG1655 carrying a non-pre-inducted pBAD_Cas9_sgRNA plasmid CamR+ (BBa_K2457006) with pRecA (AmpR+) and a linearized Donor DNA (BBa_K2457004). We screened over 94 transformants colonies, inoculating in 200µL of LB + 25µg/mL chloramphenicol + 100µg/mL ampicillin + 0.2% L-Arabinose + 1mM IPTG to activate the expression of RecA AND Cas9. We measured the initial OD (600nm) and initial fluorescence (exc. 340 nm em. 530 nm) on 96-wells plates applying the Hidex Chameleon V spectrofluorimeter. The process was repeated for each hour over 10 hours while the 96-wells plate kept growing on a shaker at 37ºC.
B) carrying a pre-inducted Cas9 + sgRNA AND RecA;
We transformed through electroporation E. coli K12 MG1655 carrying a pre-inducted pBAD_Cas9_sgRNA plasmid CamR+ (BBa_K2457006) AND pRecA plasmid AmpR+ with a linearized Donor DNA (BBa_K2457004) + pKana. We screened over 94 transformants colonies, inoculating in 200µL of LB + 50µg/mL kanamycin + 25µg/mL chloramphenicol + 100µg/mL ampicillin + 0.2% L-Arabinose + 1mM IPTG to activate the expression of RecA AND Cas9. We measured the initial OD (600nm) and initial fluorescence (exc. 340 nm em. 530 nm) on 96-wells plates applying the Hidex Chameleon V spectrofluorimeter. The process was repeated for each hour over 10 hours while the 96-wells plate kept growing on a shaker at 37ºC.
C) carrying a pre-inducted RecA.
We transformed through electroporation E. coli K12 MG1655 carrying a pRecA (AmpR+) with pBAD_Cas9_sgRNA plasmid CamR+ (BBa_K2457006) and a linearized Donor DNA (BBa_K2457004). We screened over 80 transformants colonies, inoculating in 200µL of LB + 25µg/mL chloramphenicol + 100µg/mL ampicillin + 0.2% L-Arabinose + 1mM IPTG to activate the expression of RecA AND Cas9. We measured the initial OD (600nm) and initial fluorescence (exc. 340 nm em. 530 nm) on 96-wells plates applying the Hidex Chameleon V spectrofluorimeter. The process was repeated for each hour over 10 hours while the 96-wells plate kept growing on a shaker at 37ºC.
Figure 5: CRISPeasy-in results.
Leading from the achieved results, it was noted that has certain variations between the methodologies performances: with non-induced Cas9 (30% recombinants), with pre-induced Cas9 and RecA (90% recombinants) and with exclusively pre-induced RecA (95% recombinants).
The non-induced Cas9 method possibly do not have the best performance (30%) due to the absence of Cas9 protein when the Donor DNA is inserted into genome, causing a delay in the target sequence cleavage by Cas9, and, therefore, also the delay in homologous recombination, then the major linear Donor DNA is degraded in the bacterial cytosol.
The pre-induced Cas9 and RecA presented a very good result and similar to the most effective approach. When Donor DNA is inserted into the genome, is already happening the cleavage by Cas9 and RecA is investigating a homologous sequence to the broken strand, then Donor DNA is quickly incorporated, having less DNA degradation.
We found out that the most efficient method was the one which had only RecA pre-induced, since Cas9 presence affects the cells growth, therefore, when Cas9 and Donor DNA are added simultaneously, it will have the normal cell growth, beyond the further homologous recombination.
The CRISPeasy framework development can be a boost for an easily AND reliable way to use bacterial genome editing based on CRISPR/Cas9. We encourage teams to dive into CRISPR's standardization with us and expect to be able to see in the next years more and more projects with technique's improvement!
Sequencing
Figure 6:Sequencing electropherogram from BBa_K2457004
Figure 7:Alignment of the designed sequence and our final construction from BBa_K2457004.
Sequence and Features</p>
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 321
Illegal NheI site found at 1164 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]