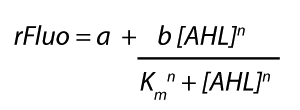Difference between revisions of "Part:BBa C0179:Experience"
(→Results) |
|||
| (15 intermediate revisions by 2 users not shown) | |||
| Line 4: | Line 4: | ||
===Applications of BBa_C0179=== | ===Applications of BBa_C0179=== | ||
| + | |||
| + | ==<b>Process measurement characterisation of LasR by Shanghaitech iGEM 2017</b>== | ||
| + | ====Group: <b>Shanghaitech 2017</b>==== | ||
| + | |||
| + | The AHL receiver LasR from <i>P.aeruginosa</i> activates expression of GFP protein in response to 3OC12-HSL. | ||
| + | |||
| + | This Receiver can be easily replaced by other AHL receivers in our collection. A full collection could be found in: http://2017.igem.org/Team:Shanghaitech/Library | ||
| + | |||
| + | |||
| + | <b>Fluorescent Response to cognate 3OC12-HSL</b> | ||
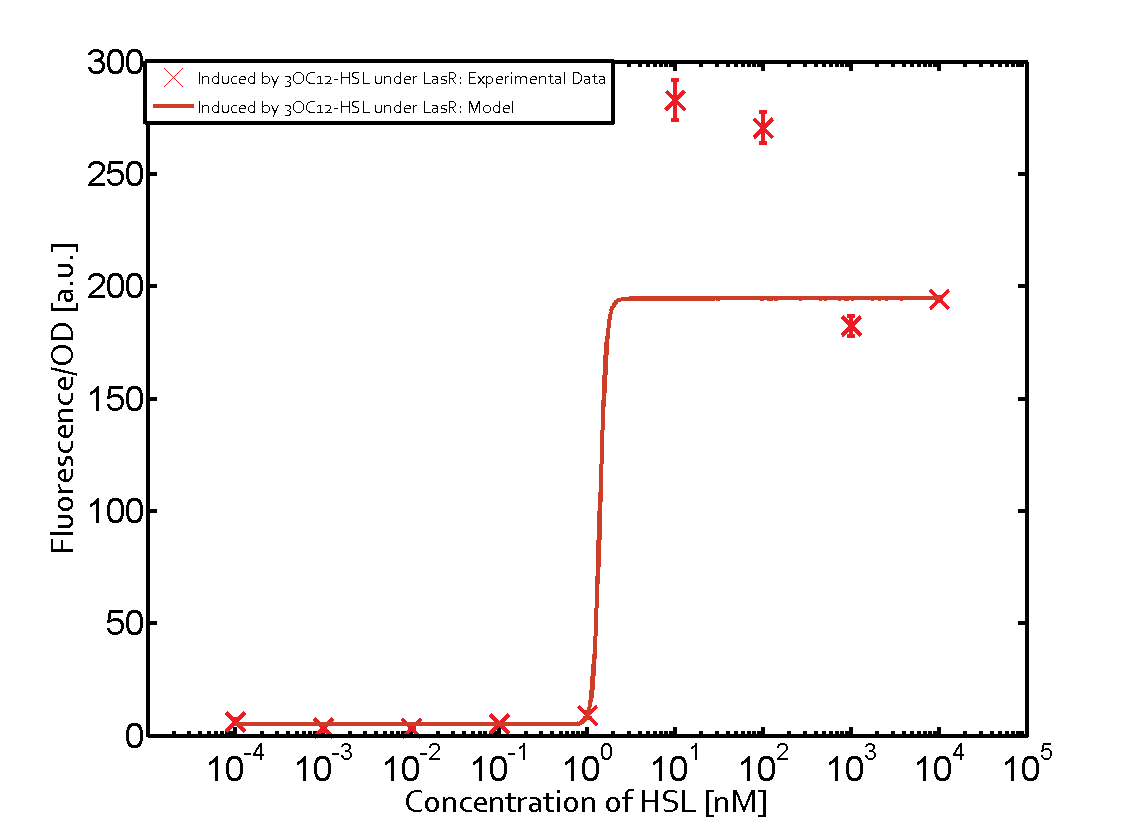
| + | *To test this part, we used standard 3OC12HSL (HSL produced by LasI in <i>P.aeruginosa</i>) to determine the response curve. | ||
| + | |||
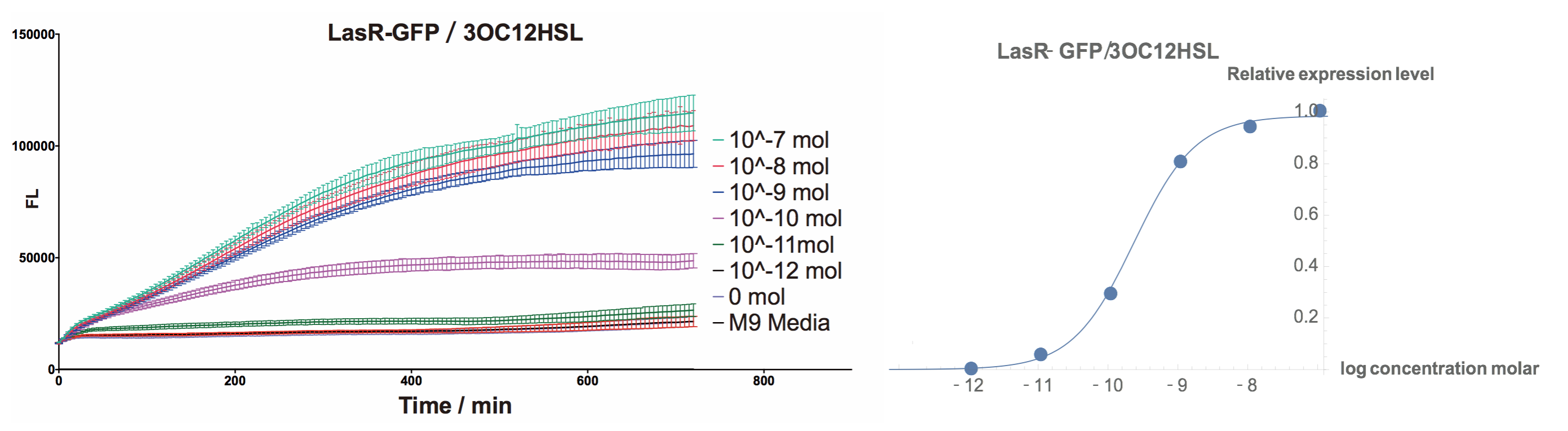
| + | [[File:T--Shanghaitech--LasRGFPLF.png|thumb|center|800px|<b>Fig. 1 LasR-pLas-GFP‘s response to cognate 3OC12HSL </b>]] | ||
| + | |||
| + | <b>Orthogonality test against non-cognate inducers</b> | ||
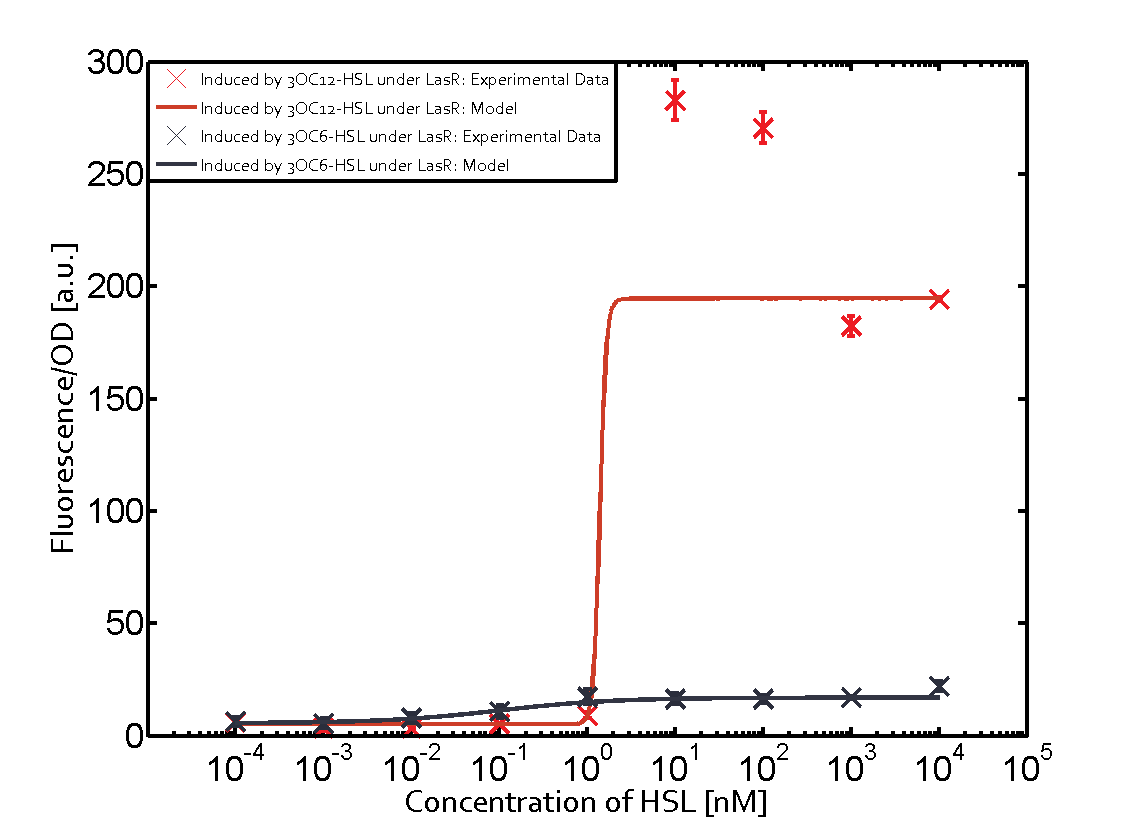
| + | *We have characterized crosstalk response of LasR to several non-cognate AHLs: | ||
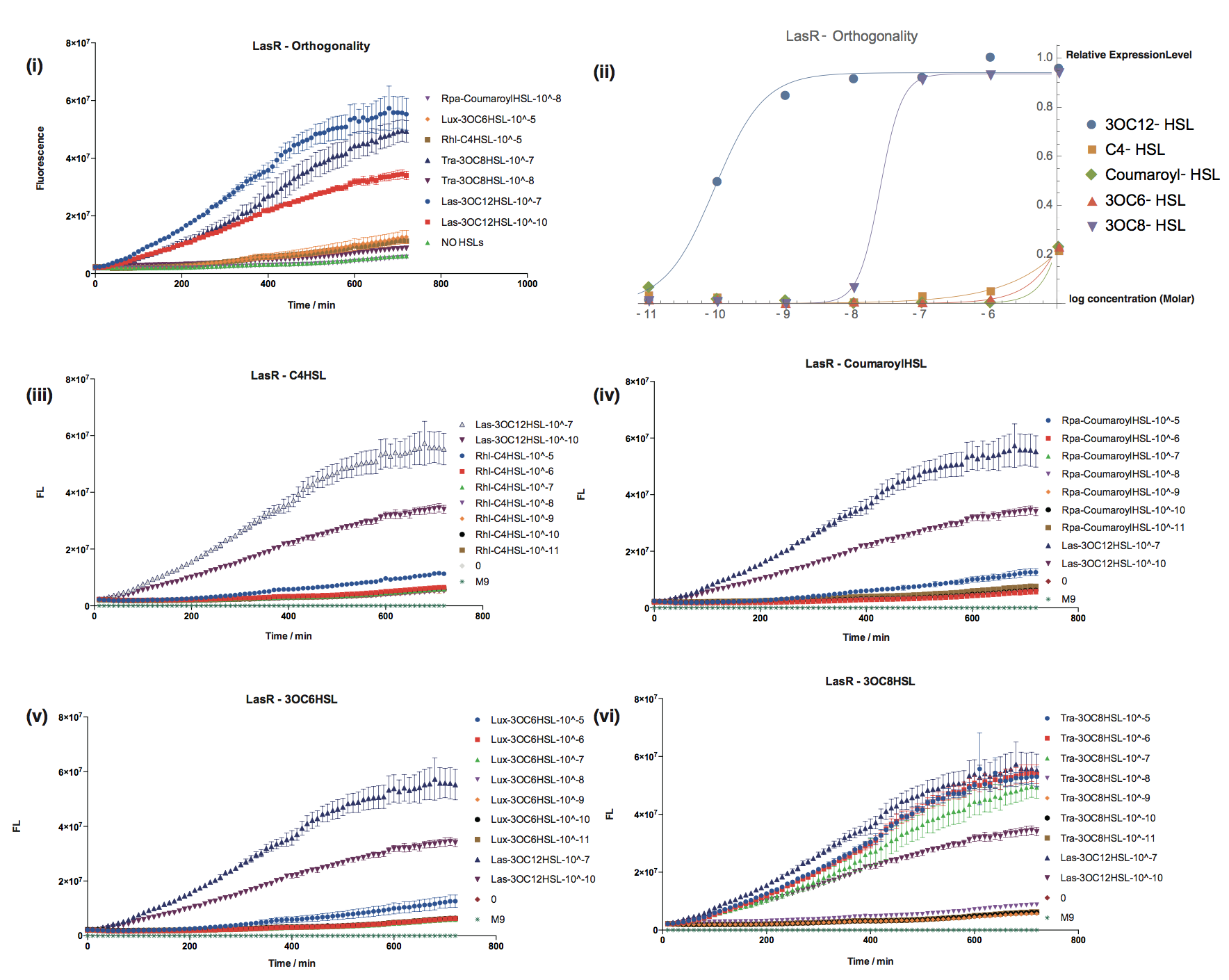
| + | [[File:T--Shanghaitech--LasRorthogonality.png|thumb|center|800px|<b>Fig. 2 Orthogonality test of LasR-pLas-GFP </b> (i):Fluorescent response to cognate and non-cognate AHLs (ii)Dose-Response curves for cognate and non-cognate AHLs (iii-vi)Fluorescent response to non-cognate AHLs in compared with 3OC12-HSL ]] | ||
| + | Furthermore, we test it under fluorescence microscope. Fig. 3 shows four testing samples. | ||
| + | [[File:T--Shanghaitech--2315034-3.png|thumb|center|700px|<b>Fig. 3 Orthogonality test under fluorescence microscope</b> <b> a) Interlab Study Test Device 1 <bbpart>BBa_J364000</bbpart> b) Las molecule 3OC12-HSL as inducer c) Tra molecule 3OC8-HSL as inducer d) Rpa molecule Coumaroyl-HSL as inducer</b>]] | ||
| + | |||
| + | It has shown that LasR is sensitive to it's cognate HSL and has obvious crosstalk with 3OC8-HSL in Tra System in relatively high concentration. | ||
| + | |||
| + | |||
| + | |||
| + | <b>Usages in our Project</b> | ||
| + | *We developed a measurement protocol using the fluorescent protein coupled AHL receiver germs to measure the actual AHLs concentration in high precision and sensitivity in compared with LC-MS. | ||
| + | |||
| + | |||
| + | |||
| + | AHL receiver from <i>P.aeruginosa</i>, actives expression of GFP protein. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
===User Reviews=== | ===User Reviews=== | ||
| Line 29: | Line 64: | ||
= Background information = | = Background information = | ||
| − | We used an ''E. coli'' TOP10 strain transformed with two medium copy plasmids (about 15 to 20 copies per plasmid and cell). The first plasmid contained the commonly used p15A origin of replication, a kanamycin resistance gene, and one out of three [https://parts.igem.org/Cell-cell_signalling cell-cell signaling promoters] ([[Part:BBa_R0062|pLux]], [[Part:BBa_R0079|pLas]], or [[Part:BBa_I14017|pRhl]]) followed by [https://parts.igem.org/Part:BBa_B0034 RBS (BBa_B0034)] and superfolder green fluorescent protein (sfGFP). In general, for spacer and terminator sequences the parts [https://parts.igem.org/Part:BBa_B0040 BBa_B0040] and [https://parts.igem.org/Part:BBa_B0015 BBa_B0015] were used, respectively. The second plasmid contained the pBR322 origin (pMB1), which yields a stable two-plasmid system together with p15A, an ampicillin resistance gene, and a [[Part:BBa_J23100|strong promoter (BBa_J23100)]] chosen from the [https://parts.igem.org/Promoters/Catalog/Anderson Anderson promoter collection] followed by [https://parts.igem.org/Part:BBa_C0179 LasR (BBa_C0179)]. The detailed construct designs and full sequences (piG0040, piG0058, piG0059, | + | We used an ''E. coli'' TOP10 strain transformed with two medium copy plasmids (about 15 to 20 copies per plasmid and cell). The first plasmid contained the commonly used p15A origin of replication, a kanamycin resistance gene, and one out of three [https://parts.igem.org/Cell-cell_signalling cell-cell signaling promoters] ([[Part:BBa_R0062|pLux]], [[Part:BBa_R0079|pLas]], or [[Part:BBa_I14017|pRhl]]) followed by [https://parts.igem.org/Part:BBa_B0034 RBS (BBa_B0034)] and superfolder green fluorescent protein (sfGFP). In general, for spacer and terminator sequences the parts [https://parts.igem.org/Part:BBa_B0040 BBa_B0040] and [https://parts.igem.org/Part:BBa_B0015 BBa_B0015] were used, respectively. The second plasmid contained the pBR322 origin (pMB1), which yields a stable two-plasmid system together with p15A, an ampicillin resistance gene, and a [[Part:BBa_J23100|strong promoter (BBa_J23100)]] chosen from the [https://parts.igem.org/Promoters/Catalog/Anderson Anderson promoter collection] followed by [https://parts.igem.org/Part:BBa_C0179 LasR (BBa_C0179)]. The detailed construct designs and full sequences (piG0040, piG0058, piG0059,piG0060) are [http://2014.igem.org/Team:ETH_Zurich/lab/sequences available here]. |
= Experimental Set-Up = | = Experimental Set-Up = | ||
| Line 43: | Line 78: | ||
== First-order crosstalk == | == First-order crosstalk == | ||
| − | In the first order crosstalk section we describe crosstalk of [https://parts.igem.org/Part:BBa_R0079 pLas] due to [https://parts.igem.org/Part:BBa_C0179 LasR] binding to inducers different from [[AHL|3OC12-HSL]] or [https://parts.igem.org/Part:BBa_R0079 pLas] itself binding to a regulator-inducer pair different from [https://parts.igem.org/Part:BBa_C0179 LasR]-[[AHL|3OC12-HSL]]. | + | In the first order crosstalk section we describe crosstalk of [https://parts.igem.org/Part:BBa_R0079 pLas] due to [https://parts.igem.org/Part:BBa_C0179 LasR] binding to inducers different from [[AHL|3OC12-HSL]], or [https://parts.igem.org/Part:BBa_R0079 pLas] itself binding to a regulator-inducer pair different from [https://parts.igem.org/Part:BBa_C0179 LasR]-[[AHL|3OC12-HSL]]. |
| − | === First Level crosstalk: LasR binds to different | + | === First Level crosstalk: LasR binds to different AHLs and activates the promoter pLas === |
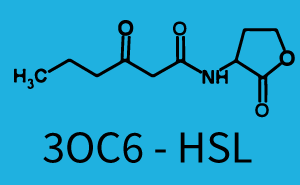
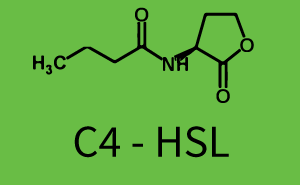
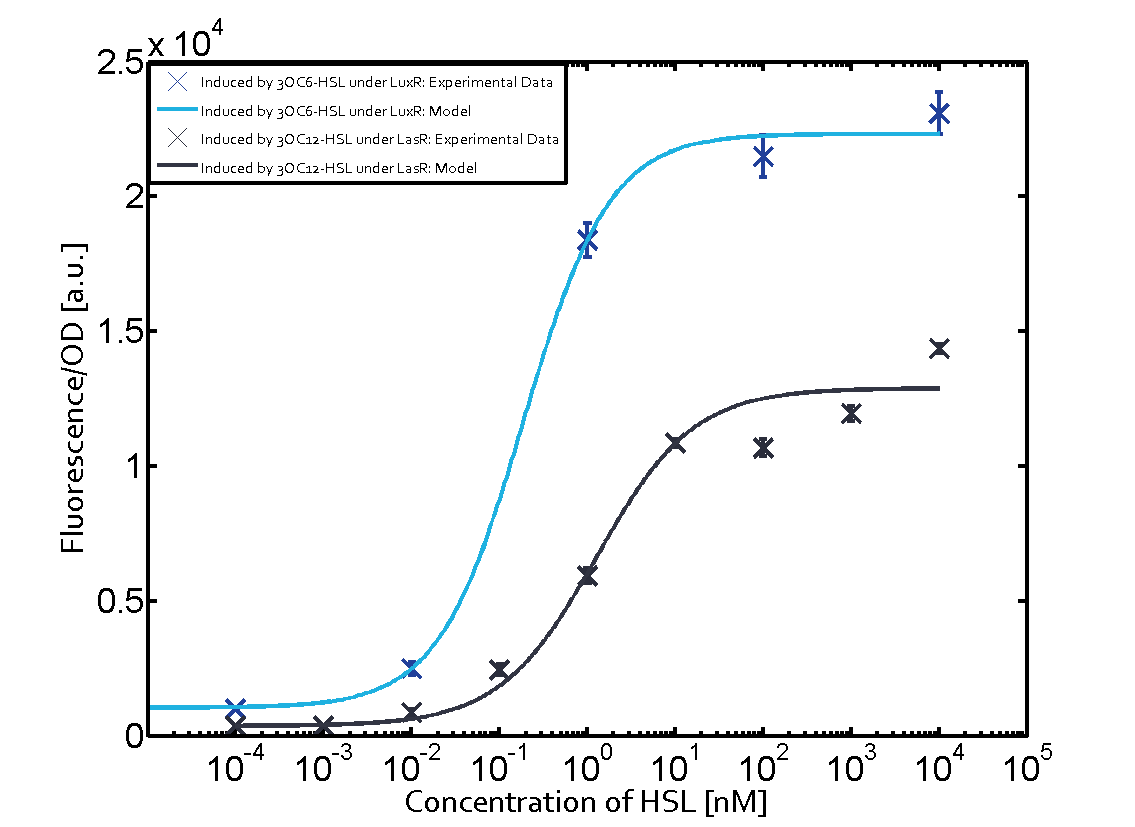
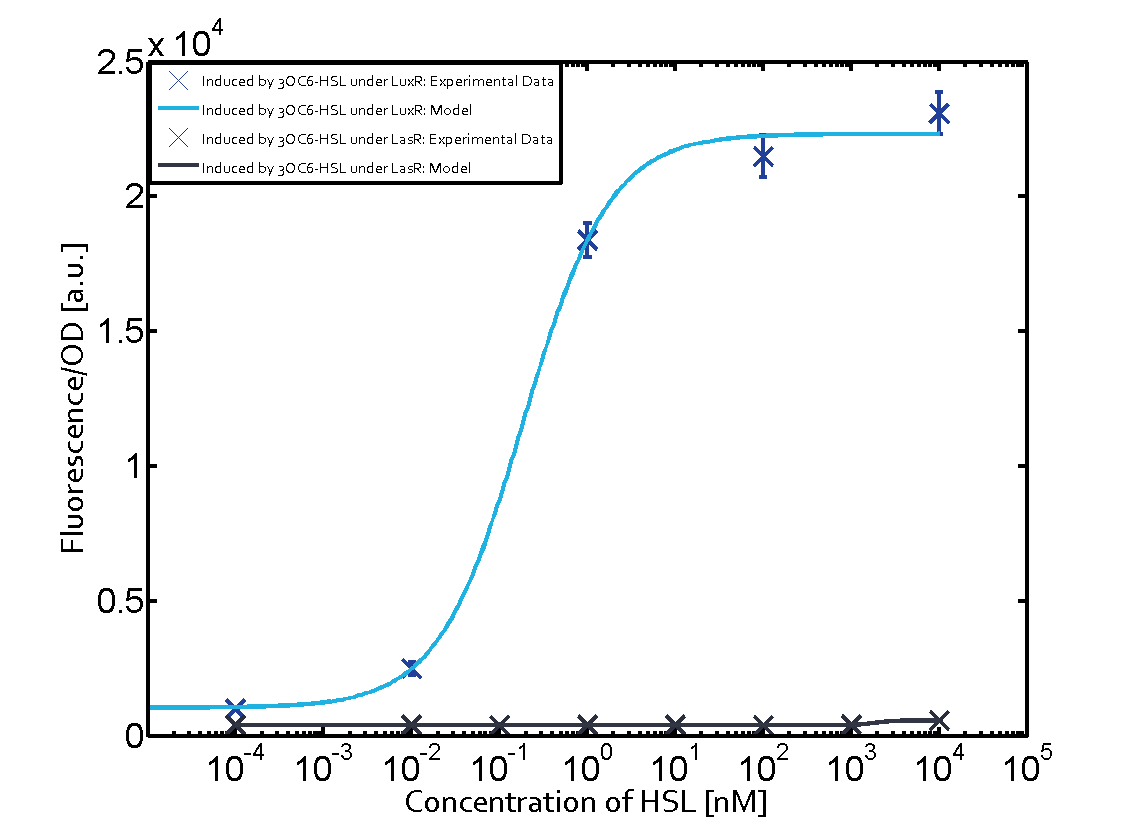
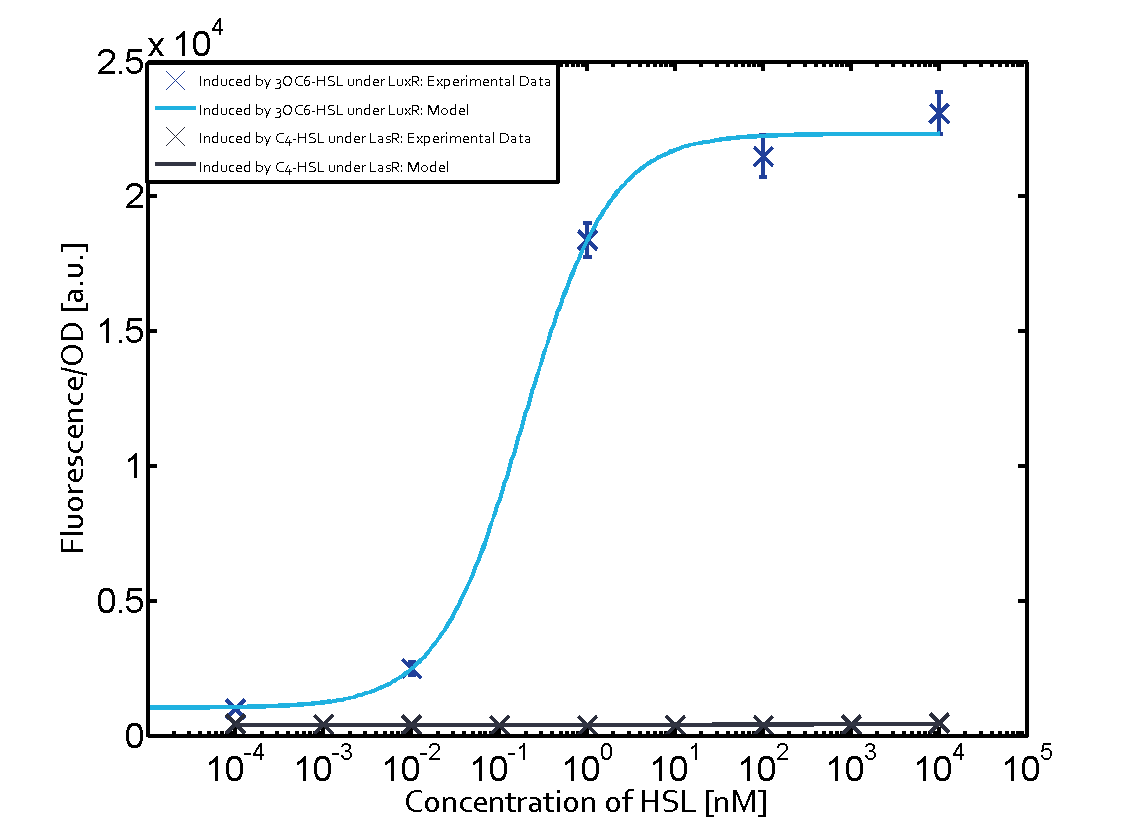
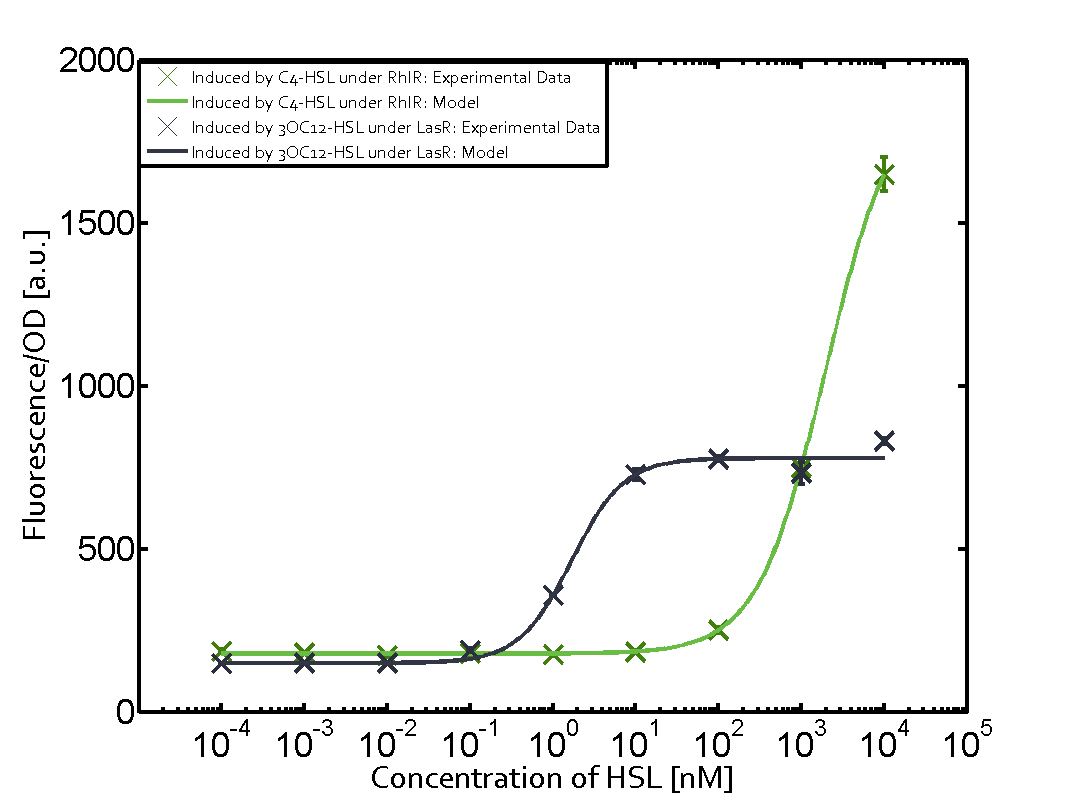
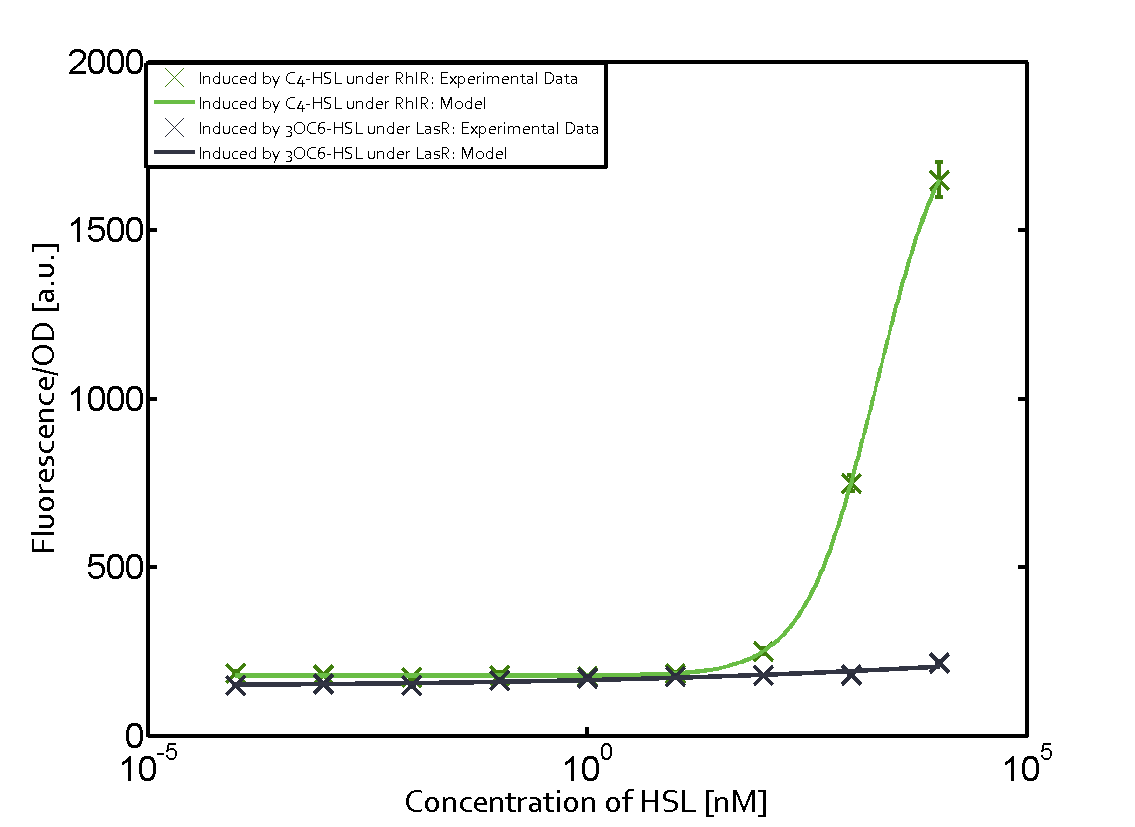
In the conventional system [[AHL|3OC12-HSL]] binds to its corresponding regulator, [https://parts.igem.org/Part:BBa_C0179 LasR], and activates the [https://parts.igem.org/Part:BBa_R0079 pLas] promoter (Figure 1, red). However, LasR can potentially also bind other AHLs and then activate pLas (Figure 1, [[3OC6HSL|3OC6-HSL]] in light blue and [[AHL|C4-HSL]] in green). | In the conventional system [[AHL|3OC12-HSL]] binds to its corresponding regulator, [https://parts.igem.org/Part:BBa_C0179 LasR], and activates the [https://parts.igem.org/Part:BBa_R0079 pLas] promoter (Figure 1, red). However, LasR can potentially also bind other AHLs and then activate pLas (Figure 1, [[3OC6HSL|3OC6-HSL]] in light blue and [[AHL|C4-HSL]] in green). | ||
| Line 55: | Line 90: | ||
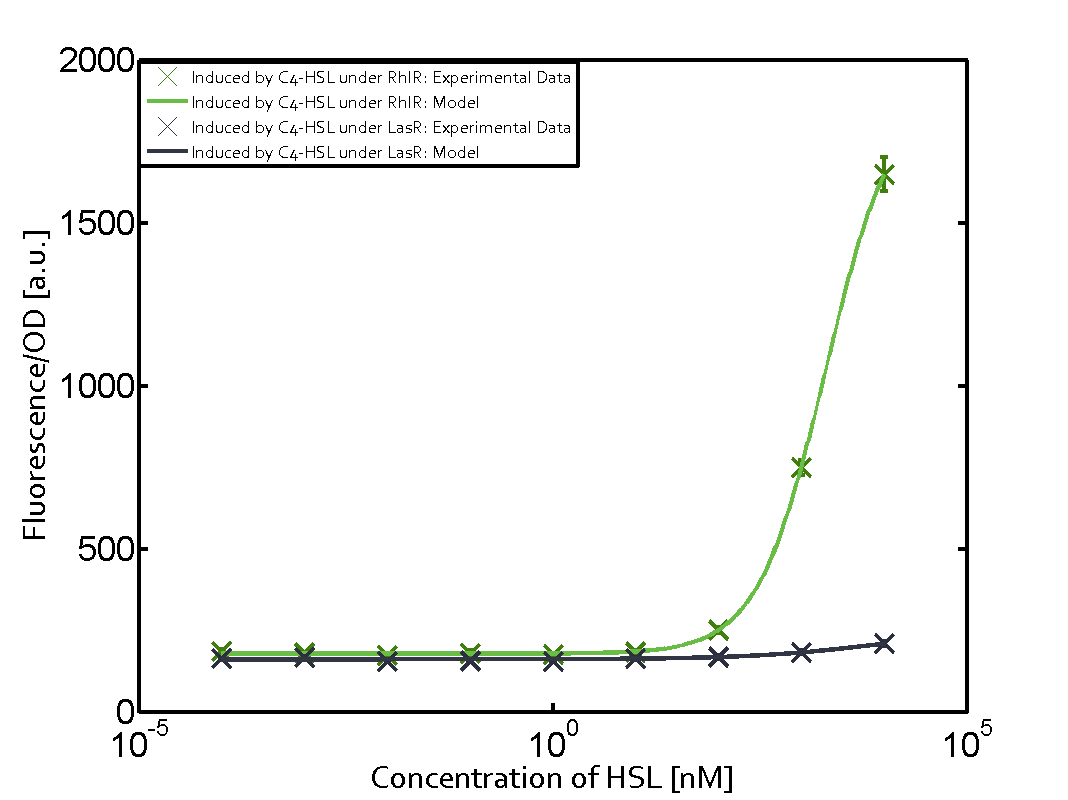
In the conventional system [[AHL|3OC12-HSL]] binds to its corresponding regulator, [https://parts.igem.org/Part:BBa_C0179 LasR], and activates the [https://parts.igem.org/Part:BBa_R0079 pLas] promoter (Figure 2 middle, red). However, [https://parts.igem.org/Part:BBa_C0179 LasR] can potentially also bind to additional [https://parts.igem.org/Cell-cell_signalling cell-cell signaling promoters] (Figure 2, [[Part:BBa_R0062|pLux]] in light blue and [[Part:BBa_I14017|pRhl]] in green) and then activate genes different from the [https://parts.igem.org/Part:BBa_R0079 pLas] regulated gene of interest . | In the conventional system [[AHL|3OC12-HSL]] binds to its corresponding regulator, [https://parts.igem.org/Part:BBa_C0179 LasR], and activates the [https://parts.igem.org/Part:BBa_R0079 pLas] promoter (Figure 2 middle, red). However, [https://parts.igem.org/Part:BBa_C0179 LasR] can potentially also bind to additional [https://parts.igem.org/Cell-cell_signalling cell-cell signaling promoters] (Figure 2, [[Part:BBa_R0062|pLux]] in light blue and [[Part:BBa_I14017|pRhl]] in green) and then activate genes different from the [https://parts.igem.org/Part:BBa_R0079 pLas] regulated gene of interest . | ||
| − | [[File:ETH Zurich | + | [[File:ETH Zurich 2014 promoter crosstalk LasR.png|400px|thumb|center| '''Figure 2 Overview of possible crosstalk of the regulator [https://parts.igem.org/Part:BBa_C0179 LasR] with three different [https://parts.igem.org/Cell-cell_signalling cell-cell signaling promoters].''' Usually, [[AHL|3OC12-HSL]] binds to its corresponding regulator, [https://parts.igem.org/Part:BBa_C0179 LasR], and activates the [https://parts.igem.org/Part:BBa_R0079 pLas] promoter (red). However, [https://parts.igem.org/Part:BBa_C0062 LasR] may also bind [[Part:BBa_R0062|pLux]] (light blue) or [[Part:BBa_I14017|pRhl]] (green) and then unintentionally activate 'off-target' gene expression.]] |
== Second order crosstalk: Combination of both cross-talk levels == | == Second order crosstalk: Combination of both cross-talk levels == | ||
| Line 61: | Line 96: | ||
The second order crosstalk describes unintended activation of a [https://parts.igem.org/Cell-cell_signalling cell-cell signaling promoter] by a mixture of both the levels described above. The regulator [https://parts.igem.org/Part:BBa_C0179 LasR] binds an inducer different from [[AHL|3OC12-HSL]] and then activates a promoter different from [https://parts.igem.org/Part:BBa_R0079 pLas]. For example, the inducer [[3OC6HSL|3OC6-HSL]] (light blue), usually binding to the regulator [https://parts.igem.org/Part:BBa_C0062 LuxR], could potentially interact with the [https://parts.igem.org/Part:BBa_C0179 LasR] regulator (red) and together activate the promoter [https://parts.igem.org/Part:BBa_I14017 pRhl] (green). This kind of crosstalk is explained in Figure 3. | The second order crosstalk describes unintended activation of a [https://parts.igem.org/Cell-cell_signalling cell-cell signaling promoter] by a mixture of both the levels described above. The regulator [https://parts.igem.org/Part:BBa_C0179 LasR] binds an inducer different from [[AHL|3OC12-HSL]] and then activates a promoter different from [https://parts.igem.org/Part:BBa_R0079 pLas]. For example, the inducer [[3OC6HSL|3OC6-HSL]] (light blue), usually binding to the regulator [https://parts.igem.org/Part:BBa_C0062 LuxR], could potentially interact with the [https://parts.igem.org/Part:BBa_C0179 LasR] regulator (red) and together activate the promoter [https://parts.igem.org/Part:BBa_I14017 pRhl] (green). This kind of crosstalk is explained in Figure 3. | ||
| − | [[File:ETH Zurich 2014 2nd order rhl.png|400px|thumb|center| '''Figure 3 Overview of possible crosstalk of the [https://parts.igem.org/Part:BBa_I14017 pRhl] promoter with both the regulator and inducer being unrelated to the promoter and each other.''' Usually, [https://parts.igem.org/Part:BBa_C0171 RhlR] together with inducer [[AHL|C4-HSL]] activate their corresponding promoter [https://parts.igem.org/Part: | + | [[File:ETH Zurich 2014 2nd order rhl.png|400px|thumb|center| '''Figure 3 Overview of possible crosstalk of the [https://parts.igem.org/Part:BBa_I14017 pRhl] promoter with both the regulator and inducer being unrelated to the promoter and each other.''' Usually, [https://parts.igem.org/Part:BBa_C0171 RhlR] together with inducer [[AHL|C4-HSL]] activate their corresponding promoter [https://parts.igem.org/Part:BBa_I14017 pRhl] (green). However, [https://parts.igem.org/Part:BBa_I14017 pRhl] may also be activated by another regulator together with an unrelated inducer. For example, the inducer [[3OC6HSL|3OC6-HSL]] (light blue) may interact with the [https://parts.igem.org/Part:BBa_C0179 LasR] regulator (red) and together activate [https://parts.igem.org/Part:BBa_I14017 pRhl] (green).]] |
== Results == | == Results == | ||
| Line 96: | Line 131: | ||
[[File:ETHZ_HillEq.png|center|200px]] | [[File:ETHZ_HillEq.png|center|200px]] | ||
<p>The fitting of the graphs was performed using the following equation :<br><br> | <p>The fitting of the graphs was performed using the following equation :<br><br> | ||
| − | rFluo = the relative fluorescence (absolute measured fluorescence value over OD)[a.u.]<br> | + | rFluo = the relative fluorescence (absolute measured fluorescence value over OD) [a.u.]<br> |
| − | a = basal expression rate [a.u.](“leakiness”)<br> | + | a = basal expression rate [a.u.] (“leakiness”)<br> |
| − | b = maximum expression rate [a.u.]("full induction")<br> | + | b = maximum expression rate [a.u.] ("full induction")<br> |
n = Hill coefficient (“cooperativity”)<br> | n = Hill coefficient (“cooperativity”)<br> | ||
K<sub>m</sub> = Half-maximal effective concentration (“sensitivity”)<br> | K<sub>m</sub> = Half-maximal effective concentration (“sensitivity”)<br> | ||
Latest revision as of 03:04, 2 November 2017
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_C0179
Process measurement characterisation of LasR by Shanghaitech iGEM 2017
Group: Shanghaitech 2017
The AHL receiver LasR from P.aeruginosa activates expression of GFP protein in response to 3OC12-HSL.
This Receiver can be easily replaced by other AHL receivers in our collection. A full collection could be found in: http://2017.igem.org/Team:Shanghaitech/Library
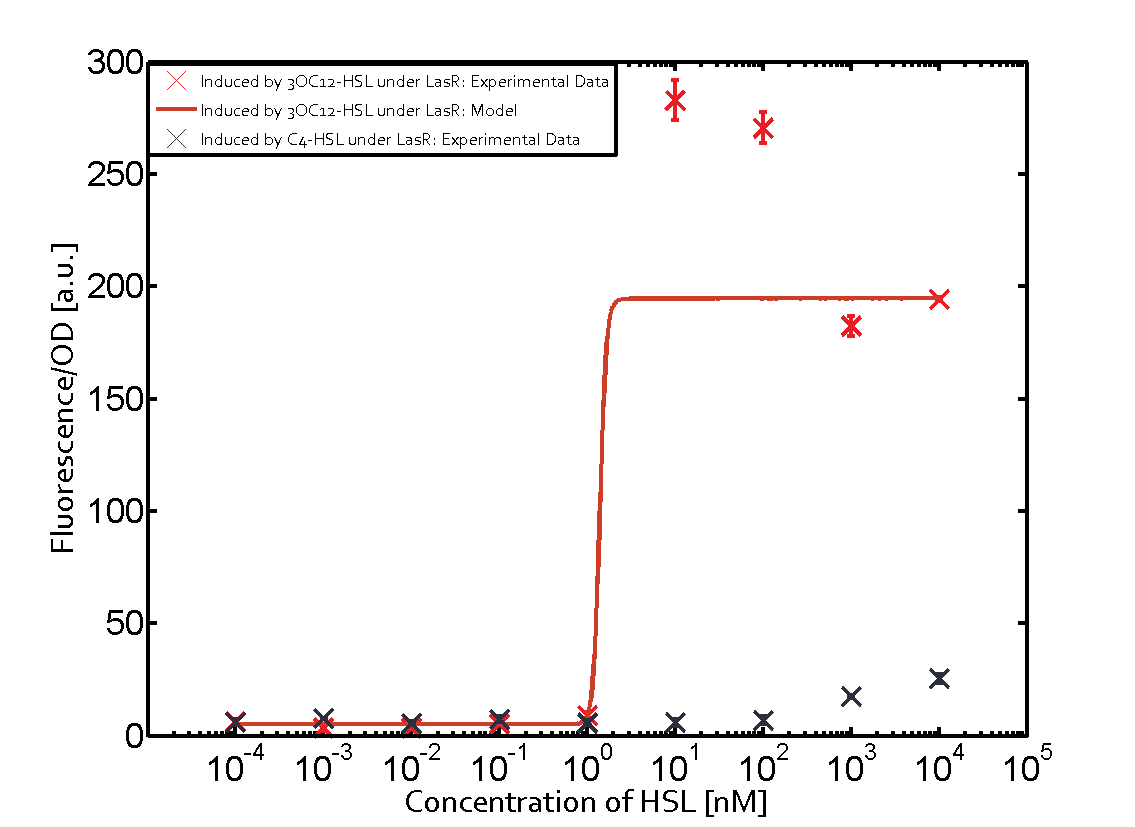
Fluorescent Response to cognate 3OC12-HSL
- To test this part, we used standard 3OC12HSL (HSL produced by LasI in P.aeruginosa) to determine the response curve.
Orthogonality test against non-cognate inducers
- We have characterized crosstalk response of LasR to several non-cognate AHLs:
Furthermore, we test it under fluorescence microscope. Fig. 3 shows four testing samples.
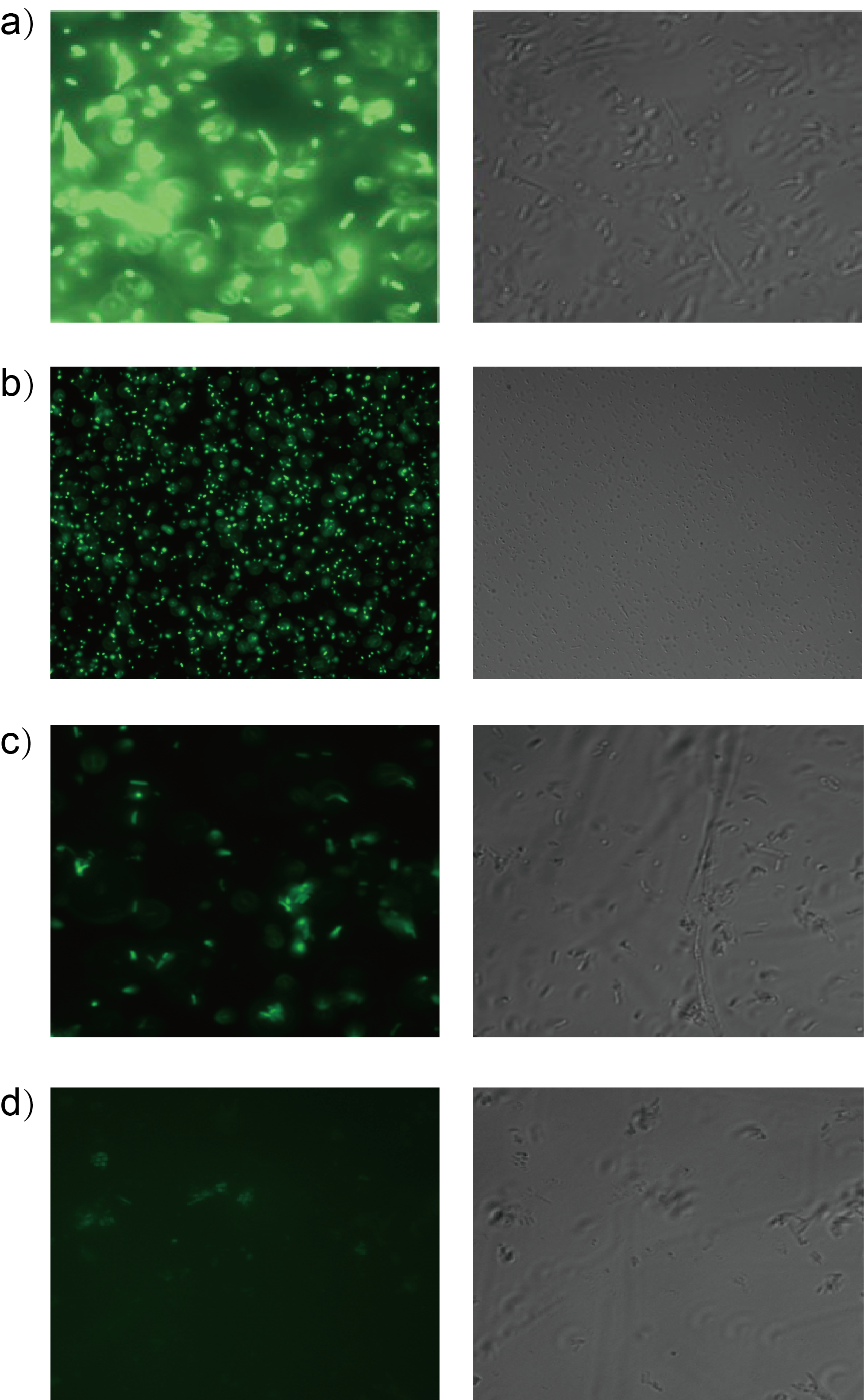
It has shown that LasR is sensitive to it's cognate HSL and has obvious crosstalk with 3OC8-HSL in Tra System in relatively high concentration.
Usages in our Project
- We developed a measurement protocol using the fluorescent protein coupled AHL receiver germs to measure the actual AHLs concentration in high precision and sensitivity in compared with LC-MS.
AHL receiver from P.aeruginosa, actives expression of GFP protein.
User Reviews
UNIQef4565567f801662-partinfo-00000001-QINU
|
••••
ETH Zurich 2014 |
Background informationWe used an E. coli TOP10 strain transformed with two medium copy plasmids (about 15 to 20 copies per plasmid and cell). The first plasmid contained the commonly used p15A origin of replication, a kanamycin resistance gene, and one out of three cell-cell signaling promoters (pLux, pLas, or pRhl) followed by RBS (BBa_B0034) and superfolder green fluorescent protein (sfGFP). In general, for spacer and terminator sequences the parts BBa_B0040 and BBa_B0015 were used, respectively. The second plasmid contained the pBR322 origin (pMB1), which yields a stable two-plasmid system together with p15A, an ampicillin resistance gene, and a strong promoter (BBa_J23100) chosen from the Anderson promoter collection followed by LasR (BBa_C0179). The detailed construct designs and full sequences (piG0040, piG0058, piG0059,piG0060) are [http://2014.igem.org/Team:ETH_Zurich/lab/sequences available here]. Experimental Set-UpThe above described E. coli TOP10 strains were grown overnight in Lysogeny Broth (LB) containing kanamycin (50 μg/mL) and ampicillin (200 μg/mL) to an OD600 of about 1.5 (37 °C, 220 rpm). As a reference, a preculture of the same strain lacking the sfGFP gene was included for each assay. The cultures were then diluted 1:40 in fresh LB containing the appropriate antibiotics and measured in triplicates in microtiter plate format on 96-well plates (200 μL culture volume) for 10 h at 37 °C with a Tecan infinite M200 PRO plate reader (optical density measured at 600 nm; fluorescence with an excitation wavelength of 488 nm and an emission wavelength of 530 nm). After 200 min we added the following concentrations of inducers (3OC6-HSL, 3OC12-HSL, and C4-HSL): 10-4 nM and 104 nM (from 100 mM stocks in DMSO). Attention: All the dilutions of 3OC12-HSL should be made in DMSO in order to avoid precipitation. In addition, in one triplicate only H2O was added as a control. From the the obtained kinetic data, we calculated mean values and plotted the dose-response-curves for 200 min past induction.
Characterization of crosstalkBackground informationHere, we focus on the characterization of crosstalk of regulator LasR and promoter pLas with different AHLs and further crosstalk of LasR and inducer C4-HSL with the three promoters - pLux, pLas, and pRhl. Also we describe a combination of the two before mentioned types of crosstalk. First-order crosstalkIn the first order crosstalk section we describe crosstalk of pLas due to LasR binding to inducers different from 3OC12-HSL, or pLas itself binding to a regulator-inducer pair different from LasR-3OC12-HSL. First Level crosstalk: LasR binds to different AHLs and activates the promoter pLasIn the conventional system 3OC12-HSL binds to its corresponding regulator, LasR, and activates the pLas promoter (Figure 1, red). However, LasR can potentially also bind other AHLs and then activate pLas (Figure 1, 3OC6-HSL in light blue and C4-HSL in green). 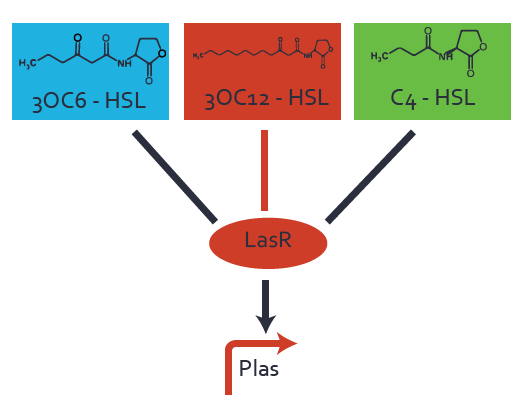 Figure 1 Overview of possible crosstalk of the LasR/pLas system with three different AHLs. Usually, 3OC12-HSL binds to its corresponding regulator, LasR, and activates the pLas promoter (red). However, LasR may also bind 3OC6-HSL (light blue) or C4-HSL (green) and then unintentionally activate pLas. Second Level crosstalk: LasR binds to 3OC12-HSL, its natural AHL, and activates promoters different from pLasIn the conventional system 3OC12-HSL binds to its corresponding regulator, LasR, and activates the pLas promoter (Figure 2 middle, red). However, LasR can potentially also bind to additional cell-cell signaling promoters (Figure 2, pLux in light blue and pRhl in green) and then activate genes different from the pLas regulated gene of interest . 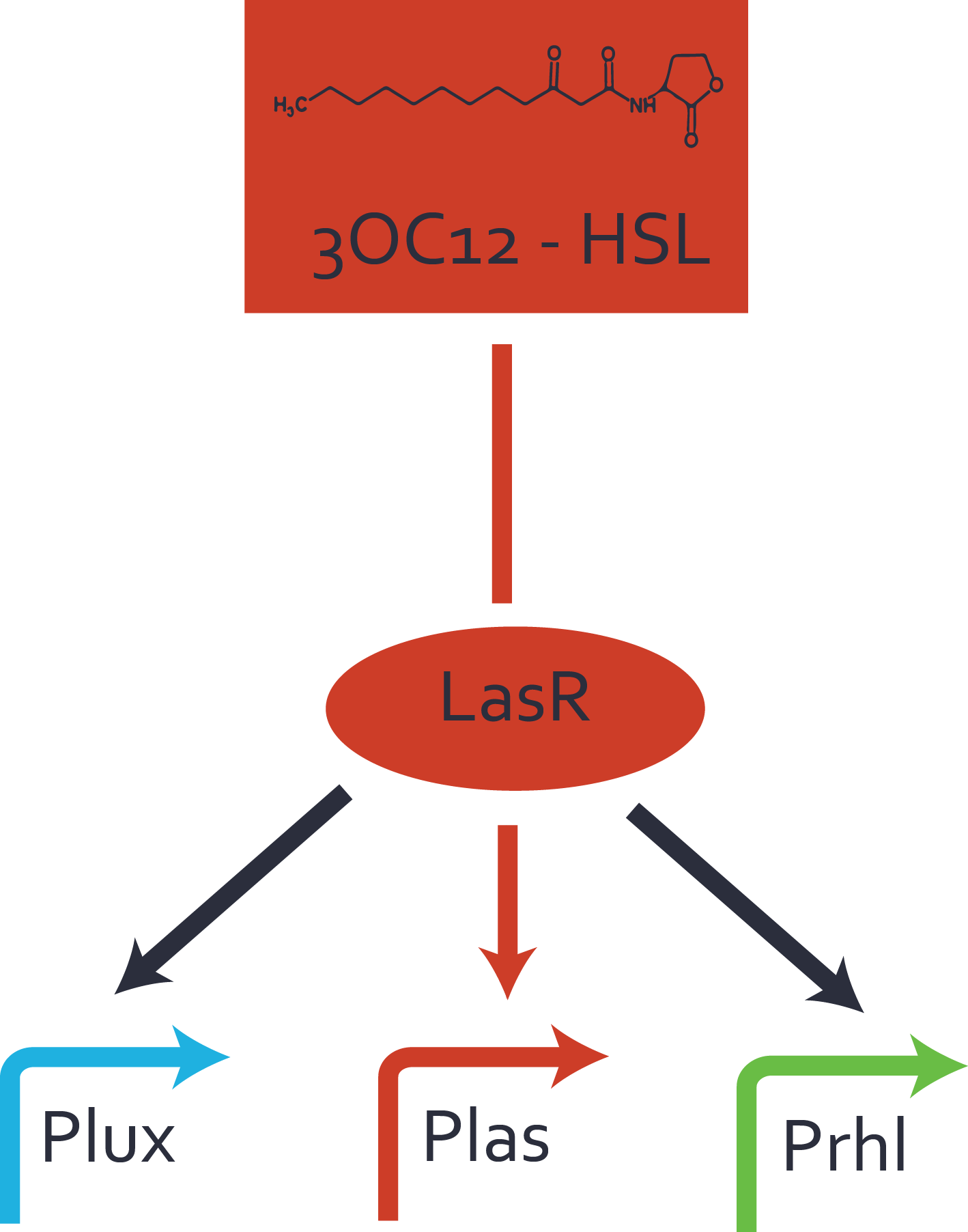 Figure 2 Overview of possible crosstalk of the regulator LasR with three different cell-cell signaling promoters. Usually, 3OC12-HSL binds to its corresponding regulator, LasR, and activates the pLas promoter (red). However, LasR may also bind pLux (light blue) or pRhl (green) and then unintentionally activate 'off-target' gene expression. Second order crosstalk: Combination of both cross-talk levelsThe second order crosstalk describes unintended activation of a cell-cell signaling promoter by a mixture of both the levels described above. The regulator LasR binds an inducer different from 3OC12-HSL and then activates a promoter different from pLas. For example, the inducer 3OC6-HSL (light blue), usually binding to the regulator LuxR, could potentially interact with the LasR regulator (red) and together activate the promoter pRhl (green). This kind of crosstalk is explained in Figure 3. 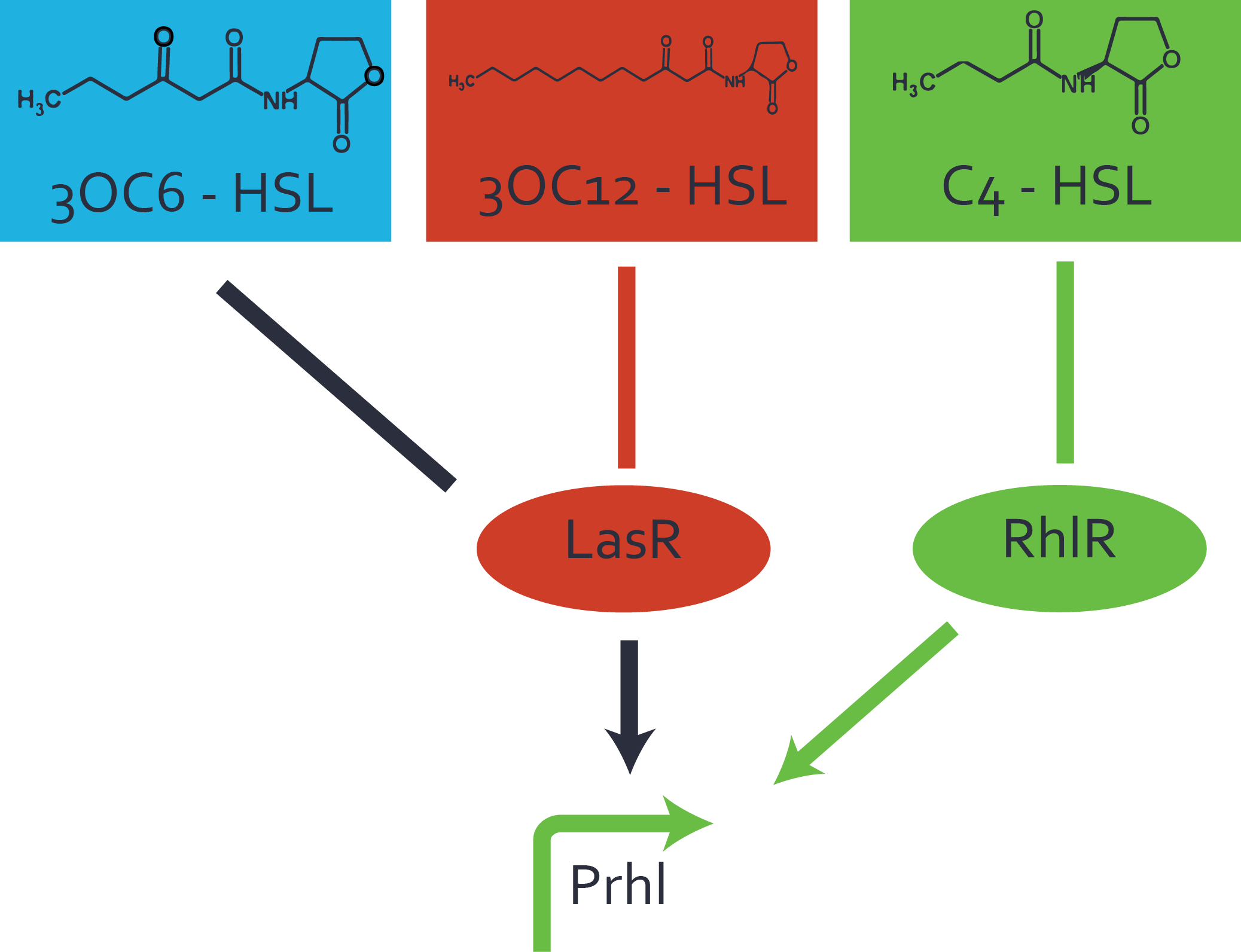 Figure 3 Overview of possible crosstalk of the pRhl promoter with both the regulator and inducer being unrelated to the promoter and each other. Usually, RhlR together with inducer C4-HSL activate their corresponding promoter pRhl (green). However, pRhl may also be activated by another regulator together with an unrelated inducer. For example, the inducer 3OC6-HSL (light blue) may interact with the LasR regulator (red) and together activate pRhl (green). Results
Modeling crosstalkEach experimental data set was fitted to an Hill function using the Least Absolute Residual method. The fitting of the graphs was performed using the following equation :
| ||||||||||||||||||||||||||||||||||||
|
Antiquity |
This review comes from the old result system and indicates that this part did not work in some test. |
|
No review score entered. Northwestern 2011 |
The 2011 Northwestern iGEM team used this part as a part of our Pseudomonas Aeruginosa biosensor. We were able to successfully express RhlR (C0171) continuously in our system. BBa K575032 |
|
•••••
iGEM Dundee 2014 |
Dundee iGEM 2014 used this lasR coding sequence to build two composite parts termed BBa_K1315009 and BBa_K1315010. These were designed as biosensors for Pseudomonas aeruginosa AutoInducer-1 (PAI-1), and were to be used in a bio-electronic device to improve diagnostics for Cystic Fibrosis patients. Details of experimental work are logged on the experience pages of BBa_K1315009 and BBa_K1315010. |