Difference between revisions of "Part:BBa K2201373"
| (50 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2201373 short</partinfo> | <partinfo>BBa_K2201373 short</partinfo> | ||
| − | T3 RNA | + | This part contains a T3 RNA Polymerase with an inverted mRFP under T3 RNA polymerase control for signal enhancing. It is an improved reporter and a genetic circuit that could report even weak expression levels. This part was designed based on the model of an amplifier in electrical engineering to intensify an existing input signal and could be used in a broad range of synthetic biology applications. We used this part for our selection system for the incorporation of non-canonical amino acids. We used this part for our selection system for the incorporation of non-canonical amino acids and demonstrate the advantages of our system in comparison with a standard reporter in an integrated modelling and wet lab characterizations. |
| − | + | <h2>Usage and Biology</h2> | |
| − | The T3 RNA | + | The T3 RNA polymerase is highly specific to T3 promoters and orthogonal to the T7 RNA polymerase. Therefore, it can be widely utilized in synthetic biology, even in expression strains which encode for a T7 RNA polymerase like <i>E. coli</i> BL21. This orthogonality is due to the promoter specifity of the different polymerases (Davis <i>et al., 1971</i>; McGraw <i>et al.</i>, 1985). |
| − | We | + | We applied this system to enhance a reporter in genetic circuits to make even weak gene expression visible. |
| − | At the moment fluorescent proteins with an emission wavelength within the visible spectra are used to report expression of the gene of interest. | + | At the moment, fluorescent proteins with an emission wavelength within the visible spectra are used to report expression of the gene of interest. Therefore, the CDS of the fluorescent protein was placed downstream of the CDS of the target protein without a terminator or promoter in between. The expression level of the target protein is nearly the same as the expression of the fluorescent protein. The fluorescence of the reporter protein indicates if the gene of interest was translated. However, thissystem is limited to strong expression, which generate a sufficienly strong fluorescence signal. |
| + | |||
| + | Therefore it is impractical for several applications which involve only weak expression. For our project, we needed a reliable and sensitive reporter to detect the expression of the gene of interest on a selection plasmid. A low expression of the target gene is essential for the selection system. No fluorescence was detectable, when the CDS of mRFP was placed downstream of the gene of interest. Through the function of the gene of interest, we knew it was expressed. To address this reporter challenge we built a genetic circuit following the model of an amplifier used in electrical engineering. | ||
| + | |||
| + | Basic amplifiers were previously submitted to the Registry of biological parts e. g. by iGEM Cambridge 2009. They build a simple circuit using an activator, which increased the transcription of a reporter under control of a second promoter. To explain their system they used the term "Polymerases per second" (PoPs). This unit defined as the flow of RNA polymerase molecules over a promoter region per second. The system developed by Cambridge 2009 (Figure 1) could increase the number of PoPs. | ||
| + | <html> | ||
| + | <div class="figure small"> | ||
| + | <img class="figure image" src="https://static.igem.org/mediawiki/2009/9/9b/Amplifier07.jpg"> | ||
| + | <p class="figure subtitle"><b>Figure 1:</b> Signal strenthening system of <a href="http://2009.igem.org/Team:Cambridge/Project/Amplification">iGEM Cambridge 2009</a>.</p> | ||
| + | </div> | ||
| + | </html> | ||
| + | To improve this system we decided to use an activator, which translates the reporter DNA and does only induce the translation of the polymerase which then translates the gene of interest. Amplification was achieved by an RNA polymerase, which transcribes the reporter DNA, multiple tmes and enhances the signal significantly.<br> | ||
| − | |||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
| Line 18: | Line 28: | ||
| − | + | <h2>Functional Parameters</h2> | |
| + | |||
| + | To design a genetic circuit that amplifies a reporter signal we decided to use an RNA polymerase, orthogonal to the native <i>E. coli</i> RNA polymerase. Therefore, the T3 RNA polymerase previously characterized by iGEM Peking 2010 was deployed. The T3 RNA polymerase is highly specific to T3 promoters and orthogonal to the <i>E. coli </i>DNA polymerases and even to the T7 RNA polymerase. We decided not to use the T7 RNA polymerase, because it is already part of expression strains like BL21 and would therefore prevent the application in such strains. The construction of our designed composite part comprising a standard mRFP reporter is shown in Figure 2. | ||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--SVI-composite-part.png|800px|thumb|center|<b>Figure 2:</b> Construction of the standard mRFP reporter and the genetic circuit for the amplification of mRFP expression. In construct 1 the CDS for the mRFP transcript is downstream of the CDS of the gene of interest. In construct 2 the CDS of the mRFP is downstream a T3 promoter and the CDS coding for the T3 RNA polymerase is downstream the CDS of the gene of interest.]]<br> | ||
| + | |||
| + | Figure 2 shows two reporter constructs for gene expression quantification. The first one is the standard reporter for the gene expression of the target gene, using mRFP as reporter gene. If the gene of interest is expressed, the mRFP is expressed at nearly the same level. The mRFP fluorescence reveals the gene expression of the gene of interest. This system is suitable for but also limited to high expression rates. Low, expression of the target gene is associated with a low expression of mRFP and thus not visible. | ||
| + | |||
| + | The second system is our genetic circuit consisting of a CDS of the target gene upstream of the CDS for a T3 DNA polymerase. Therefore, the expression of the reporter gene is nearly on the same level as the expression of the target gene. The expressed T3 RNA polymerase transcribes the mRFP under the control of the T3 promoter. | ||
| + | |||
| + | |||
| + | <h2>Modeling</h2> | ||
| + | |||
| + | To demonstrate the advantages of our improved construct, we modeled the amount of mRFP transcript for both constructs. | ||
| + | If we assume the expression of the gene of interest is low and only one <i>E. coli</i> RNA polymerase with a chain elongation rate of 50 nucleotides per second translates the both products, construct 1 produces 1 mRFP transcript in 16 seconds. Construct 2 expresses 1 T3 RNA polymerase transcript every 52 seconds. After translation (with an average translation rate of 20 amino acids per second ~42 sec), these polymerases transcribe the mRFP transcript with a chain elongation rate of 170 nucleotides per second. Therefore, every T3 RNA polymerase generates one mRFP transcript every 4.7 seconds. The resulting amount of mRFP transcripts is shown in Figure 3. The script for our modelling can be found <html><a href="https://static.igem.org/mediawiki/parts/8/8a/T--Bielefeld-CeBiTec--model-composite-2.txt">here</a></html>. | ||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--model-composite.png|600px|thumb|center|<b>Figure 3:</b> Modeling on the amount of mRFP transcript transcribed through the two different models. In model 1 the CDS fot the mRFP transcript is downstream the CDS of the gene of interest. In model 2 the CDS of the mRFP is downstream a T3 promoter and the CDS coding for the T3 RNA polymerase is downstream the CDS of the gene of interest.]]<br> | ||
| + | |||
| + | The number of mRFP transcripts is a lot higher with our signal enhancement system (construct 2) even through this model leaves out the fact that one transcript of the T3 RNA polymerase could be translated several times and enhance the signal even more. Despite the simplicity of this model, it shows the advantages of using the T3 RNA polymerase for signal enhancing. | ||
| + | |||
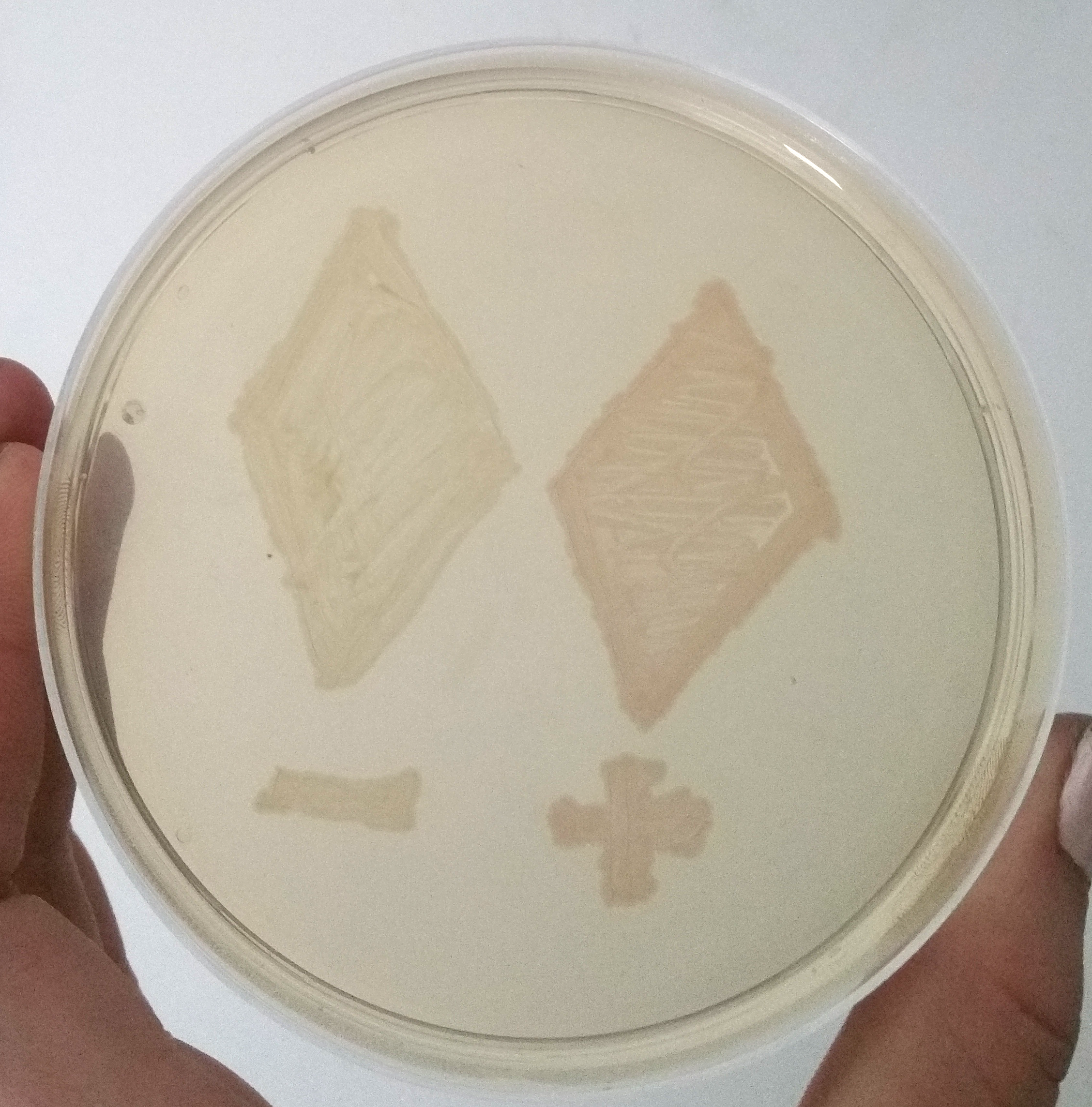
| + | To demonstrate the signal enhancing system we cloned either the part <html><a href="https://parts.igem.org/Part:BBa_E1010">E1010</a></html> or our signal enhancing system <html> <a href="https://parts.igem.org/Part:BBa_K2201373">(BBa_K2201373)</a></html> downstream of the part <html><a href="https://parts.igem.org/Part:BBa_K2201900">BBa_K2201900</a></html>. This plasmid was characterized in cells containing our positive selection plasmid. A smear of the transformants is shown in Figure 4. | ||
| + | |||
| + | [[File:T--Bielefeld-CeBiTec--composite-1.png|600px|thumb|center|<b>Figure 4:</b> Two Smear of two clones containing only mRFP under an uninduced T7 promoter (-) and containing the mRFP enhancing system under the same promotor (+), after 12 h of incubation at 37 °C.]]<br> | ||
| − | + | The uninduced T7 promoter leads only to a basal transcription level. Despite this weak transcription, the mRFP in the cells containing the signal enhancing system is clearly visible, while the cells without the signal enhancement system remain colorless, like assumed in our modeling. | |
| − | + | <h2>Usage</h2> | |
| + | For the selection of tRNA/aminoacylsynthetase to incorporate non-canonical amino acids, we needed to check if clones still contain the positive selection plasmid in the negative selection round. Therefore, we decided to incorporate the mRFP signal enhancing system downstream of the CDS of our positive selection plasmid <html><a href="https://parts.igem.org/Part:BBa_K2201900">BBa_K2201900</a></html>. If the cells contain this plasmid in the negative selection, they should be visible as red clones. In contrast, the clones containing the negative selection plasmid should be colorless. A picture of one round of the negative selection (Figure 5) shows that one clone is red, thus demonstrating the function of the system. | ||
| − | Figure | + | [[File:T--Bielefeld-CeBiTec--composite-2.png|600px|thumb|center|<b>Figure 5:</b> Picture of a negative selection round. The clon still containing the positive selection plasmid, thus the mRFP enhancing system, is red.]]<br> |
| − | |||
| + | More informations and the reults of our selection system for the incorporation of non-canonical amino acids can be found <html><a href="http://2017.igem.org/Team:Bielefeld-CeBiTec/Results/translational_system/library_and_selection">here</a></html>. | ||
| + | <h2>Applications</h2> | ||
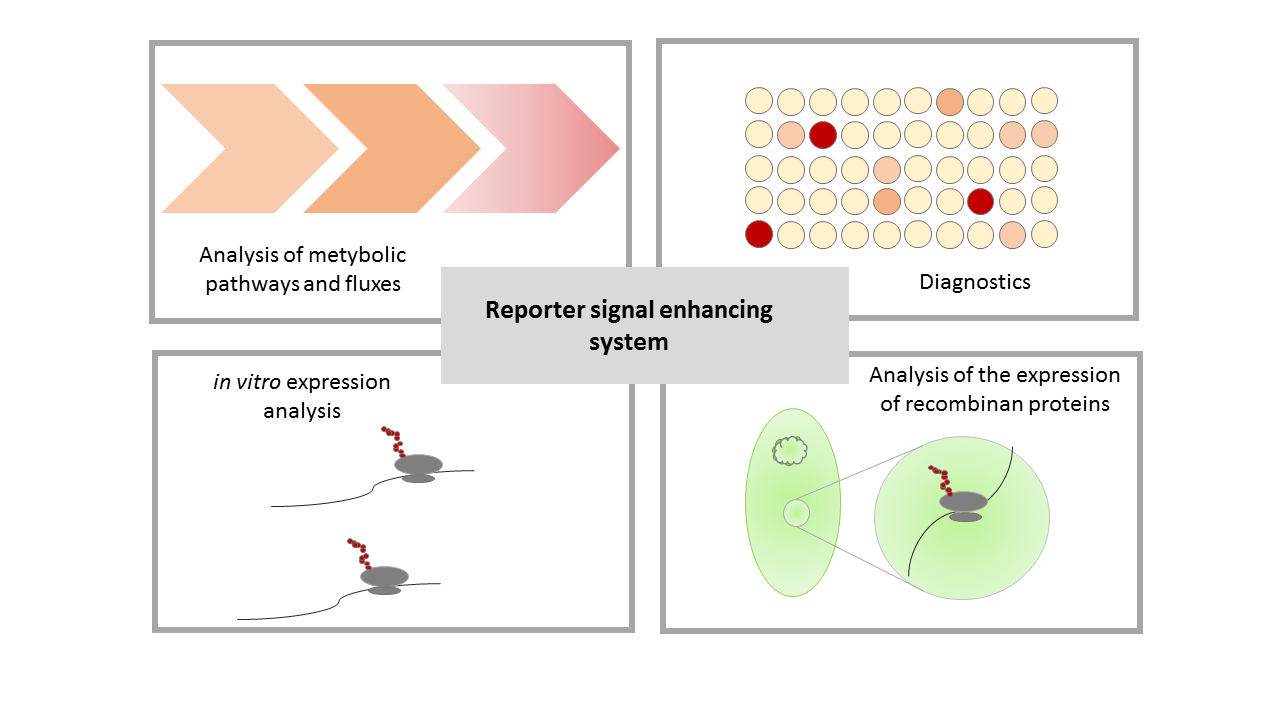
| + | An important criteria for the best composite part is the possibility to use this part in further iGEM projects. In addition to our application for this part, there are a lot of potential applications for a reporter signal enhancement. Our system provides a reliable and especially sensitive reporter, thus applications which require these could be improved by using our system. Some possible applications are shown in Figure 6. | ||
| + | <div class="figure large"> | ||
| + | [[File:T--Bielefeld-CeBiTec--composite-6.png|800px|thumb|center|<b>Figure 6: Potential applications for the reporter signal enhancing system.</b> The sensitivity and high output signal enable could improve projects for metabolic and fluxom analysis, diagnostics, <i>in vitro</i> expression analysisi and detection of recombinant expression under weak promoters. ]]<br> | ||
| + | |||
| + | The strength of the reporter signal enhancing system lies in its strong and specific signal despite of a very weak initial gene expression. This enables usage in the analysis of weakly transcribed genes in metabolic pathway and flux analyses as well as a variety of diagnostic application which are dependent on strong, specific signals. Furthermore use of the reporter signal enhancer system in the analysis of expression in vitro and in vivo is feasible. | ||
| + | <br> | ||
| + | <h2>References</h2> | ||
| + | <b>Davis, R. W. & Hyman, R. W </b>(1971)J. Mol. Biol <b>62</b>, 287-301. <br> | ||
| + | <b>McGraw, N. J., Bailey, J. N., Cleaves, G. R., Dembinski, D. R., Gocke, C. R., Joliffe, L. K., MacWright, R. S.&McAllister, W. T.</b> (1985) Nucleic Acids Res. <b>13</b>, 6753–6766. | ||
<partinfo>BBa_K2201373 parameters</partinfo> | <partinfo>BBa_K2201373 parameters</partinfo> | ||
Latest revision as of 01:14, 2 November 2017
T3 polymerase with inverted mRFP under T3 promoter control for signal enhancing
This part contains a T3 RNA Polymerase with an inverted mRFP under T3 RNA polymerase control for signal enhancing. It is an improved reporter and a genetic circuit that could report even weak expression levels. This part was designed based on the model of an amplifier in electrical engineering to intensify an existing input signal and could be used in a broad range of synthetic biology applications. We used this part for our selection system for the incorporation of non-canonical amino acids. We used this part for our selection system for the incorporation of non-canonical amino acids and demonstrate the advantages of our system in comparison with a standard reporter in an integrated modelling and wet lab characterizations.
Usage and Biology
The T3 RNA polymerase is highly specific to T3 promoters and orthogonal to the T7 RNA polymerase. Therefore, it can be widely utilized in synthetic biology, even in expression strains which encode for a T7 RNA polymerase like E. coli BL21. This orthogonality is due to the promoter specifity of the different polymerases (Davis et al., 1971; McGraw et al., 1985). We applied this system to enhance a reporter in genetic circuits to make even weak gene expression visible.
At the moment, fluorescent proteins with an emission wavelength within the visible spectra are used to report expression of the gene of interest. Therefore, the CDS of the fluorescent protein was placed downstream of the CDS of the target protein without a terminator or promoter in between. The expression level of the target protein is nearly the same as the expression of the fluorescent protein. The fluorescence of the reporter protein indicates if the gene of interest was translated. However, thissystem is limited to strong expression, which generate a sufficienly strong fluorescence signal.
Therefore it is impractical for several applications which involve only weak expression. For our project, we needed a reliable and sensitive reporter to detect the expression of the gene of interest on a selection plasmid. A low expression of the target gene is essential for the selection system. No fluorescence was detectable, when the CDS of mRFP was placed downstream of the gene of interest. Through the function of the gene of interest, we knew it was expressed. To address this reporter challenge we built a genetic circuit following the model of an amplifier used in electrical engineering.
Basic amplifiers were previously submitted to the Registry of biological parts e. g. by iGEM Cambridge 2009. They build a simple circuit using an activator, which increased the transcription of a reporter under control of a second promoter. To explain their system they used the term "Polymerases per second" (PoPs). This unit defined as the flow of RNA polymerase molecules over a promoter region per second. The system developed by Cambridge 2009 (Figure 1) could increase the number of PoPs.

Figure 1: Signal strenthening system of iGEM Cambridge 2009.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 2717
Illegal AgeI site found at 2829 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 2585
Functional Parameters
To design a genetic circuit that amplifies a reporter signal we decided to use an RNA polymerase, orthogonal to the native E. coli RNA polymerase. Therefore, the T3 RNA polymerase previously characterized by iGEM Peking 2010 was deployed. The T3 RNA polymerase is highly specific to T3 promoters and orthogonal to the E. coli DNA polymerases and even to the T7 RNA polymerase. We decided not to use the T7 RNA polymerase, because it is already part of expression strains like BL21 and would therefore prevent the application in such strains. The construction of our designed composite part comprising a standard mRFP reporter is shown in Figure 2.
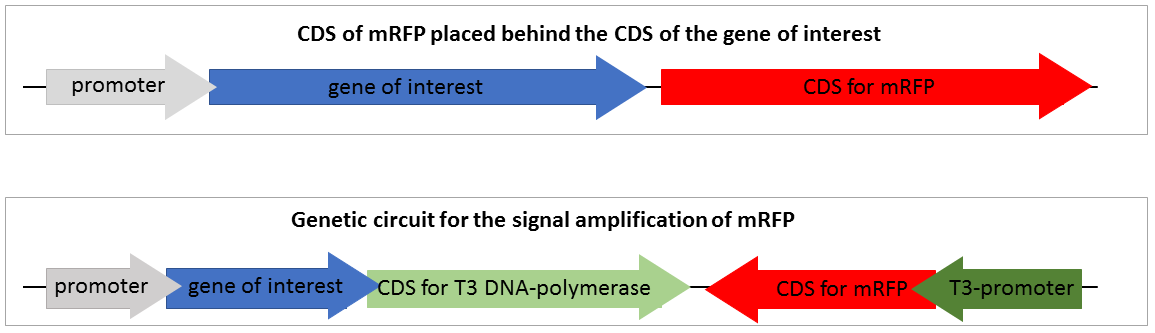
Figure 2 shows two reporter constructs for gene expression quantification. The first one is the standard reporter for the gene expression of the target gene, using mRFP as reporter gene. If the gene of interest is expressed, the mRFP is expressed at nearly the same level. The mRFP fluorescence reveals the gene expression of the gene of interest. This system is suitable for but also limited to high expression rates. Low, expression of the target gene is associated with a low expression of mRFP and thus not visible.
The second system is our genetic circuit consisting of a CDS of the target gene upstream of the CDS for a T3 DNA polymerase. Therefore, the expression of the reporter gene is nearly on the same level as the expression of the target gene. The expressed T3 RNA polymerase transcribes the mRFP under the control of the T3 promoter.
Modeling
To demonstrate the advantages of our improved construct, we modeled the amount of mRFP transcript for both constructs. If we assume the expression of the gene of interest is low and only one E. coli RNA polymerase with a chain elongation rate of 50 nucleotides per second translates the both products, construct 1 produces 1 mRFP transcript in 16 seconds. Construct 2 expresses 1 T3 RNA polymerase transcript every 52 seconds. After translation (with an average translation rate of 20 amino acids per second ~42 sec), these polymerases transcribe the mRFP transcript with a chain elongation rate of 170 nucleotides per second. Therefore, every T3 RNA polymerase generates one mRFP transcript every 4.7 seconds. The resulting amount of mRFP transcripts is shown in Figure 3. The script for our modelling can be found here.
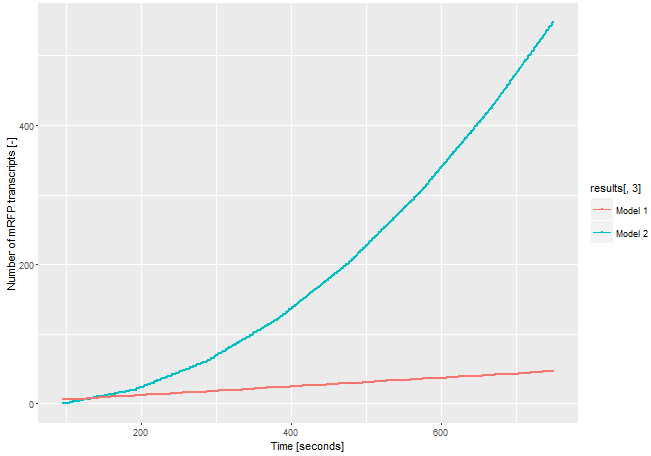
The number of mRFP transcripts is a lot higher with our signal enhancement system (construct 2) even through this model leaves out the fact that one transcript of the T3 RNA polymerase could be translated several times and enhance the signal even more. Despite the simplicity of this model, it shows the advantages of using the T3 RNA polymerase for signal enhancing.
To demonstrate the signal enhancing system we cloned either the part E1010 or our signal enhancing system (BBa_K2201373) downstream of the part BBa_K2201900. This plasmid was characterized in cells containing our positive selection plasmid. A smear of the transformants is shown in Figure 4.
The uninduced T7 promoter leads only to a basal transcription level. Despite this weak transcription, the mRFP in the cells containing the signal enhancing system is clearly visible, while the cells without the signal enhancement system remain colorless, like assumed in our modeling.
Usage
For the selection of tRNA/aminoacylsynthetase to incorporate non-canonical amino acids, we needed to check if clones still contain the positive selection plasmid in the negative selection round. Therefore, we decided to incorporate the mRFP signal enhancing system downstream of the CDS of our positive selection plasmid BBa_K2201900. If the cells contain this plasmid in the negative selection, they should be visible as red clones. In contrast, the clones containing the negative selection plasmid should be colorless. A picture of one round of the negative selection (Figure 5) shows that one clone is red, thus demonstrating the function of the system.
More informations and the reults of our selection system for the incorporation of non-canonical amino acids can be found here.
Applications
An important criteria for the best composite part is the possibility to use this part in further iGEM projects. In addition to our application for this part, there are a lot of potential applications for a reporter signal enhancement. Our system provides a reliable and especially sensitive reporter, thus applications which require these could be improved by using our system. Some possible applications are shown in Figure 6.
The strength of the reporter signal enhancing system lies in its strong and specific signal despite of a very weak initial gene expression. This enables usage in the analysis of weakly transcribed genes in metabolic pathway and flux analyses as well as a variety of diagnostic application which are dependent on strong, specific signals. Furthermore use of the reporter signal enhancer system in the analysis of expression in vitro and in vivo is feasible.
References
Davis, R. W. & Hyman, R. W (1971)J. Mol. Biol 62, 287-301.
McGraw, N. J., Bailey, J. N., Cleaves, G. R., Dembinski, D. R., Gocke, C. R., Joliffe, L. K., MacWright, R. S.&McAllister, W. T. (1985) Nucleic Acids Res. 13, 6753–6766.