Difference between revisions of "Part:BBa K1998012"
SWinchester (Talk | contribs) |
Ari edmonds (Talk | contribs) |
||
| (9 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1998012 short</partinfo> | <partinfo>BBa_K1998012 short</partinfo> | ||
| + | |||
| + | The wrong sequence was submitted to this page in 2016. iGEM Macquarie 2017 has created a new updated part page with the correct sequence (BBa_K2300003) (https://parts.igem.org/Part:BBa_K2300003#Overview). | ||
<!-- --> | <!-- --> | ||
| Line 15: | Line 17: | ||
The HydEF protein contains two unique domains that are homologous to two distinct prokaryotic proteins, HydE and HydF, which are found exclusively in organisms containing [Fe] hydrogenase. This part makes up one of the parts found in an operon of the hydrogen production pathway. | The HydEF protein contains two unique domains that are homologous to two distinct prokaryotic proteins, HydE and HydF, which are found exclusively in organisms containing [Fe] hydrogenase. This part makes up one of the parts found in an operon of the hydrogen production pathway. | ||
<br><br> | <br><br> | ||
| − | |||
| − | <html><center><img src="https://static.igem.org/mediawiki/ | + | <html><center><img src="https://static.igem.org/mediawiki/parts/b/b5/HydrogenPathwayUpdated2016.jpeg" alt="HydrogenProduction" height="50%"width="75%"></center></html> |
===Biology & Literature=== | ===Biology & Literature=== | ||
| − | + | <i>hydEF</i> (comprising of <i>HydE</i> and <i>HydF</i>) encodes proteins necessary for hydrogenase activity by functioning in H cluster biosynthesis and enzyme maturation involving S-adenosylmethionine (SAM) dependent mechanisms [1]. <i>HydF</i> is part of the GTPase protein family, with GTP-binding motifs present at the N terminal end and iron-sulfur cluster-binding motifs at the C terminal end [2]. Both <i>HydE</i> is a radical SAM (S-adenosyl methionine) enzyme [3]. King et al. stated that challenges faced during the expression of maturation genes <i>HydEF</i> and <i>HydG</i> in E. coli were that they were unstable and prone to rearrangements [1]. | |
<br> | <br> | ||
<br> | <br> | ||
| − | Both | + | Both <i>HydEF</i> and <i>HydG</i> are transcribed under anaerobic conditions and are necessary for the assembly of an active <i>HydA1</i> hydrogenase [2]. The importance of <i>HydEF</i> in hydrogenase activity has also been illustrated in the mutant <i>hydEF-1</i> strain of <i>C. reinhardtii</i> which is unable to produce H2 due to a non-functional HydEF gene [4]. |
<br> | <br> | ||
| + | ===Part Verification=== | ||
| + | |||
| + | <html><center><img src="https://static.igem.org/mediawiki/2016/b/bd/T--Macquarie_Australia--HYDShowGel.png" " width="50%" height="35%"></center> </html> | ||
| + | <b>Fig 1.</b> A gel consisting of EcorI/PstI for the hydEF part. The expected band for the hydEF (3611 bp) part was observed. | ||
===Protein information=== | ===Protein information=== | ||
| − | + | <i>HydEF</i><br> | |
| + | Mass: 121.95 kDa<br> | ||
| + | Sequence: <br> | ||
MAHSLSAHSRQAGDRKLGAGAASSRPSCPSRRIVRVAAHASASKATPDVPVDDLPPAHARAAVAAANRRARAMASAEAAAETLGDFLGLGKGGLSP | MAHSLSAHSRQAGDRKLGAGAASSRPSCPSRRIVRVAAHASASKATPDVPVDDLPPAHARAAVAAANRRARAMASAEAAAETLGDFLGLGKGGLSP | ||
GATANLDREQVLGVLEAVWRRGDLNLERALYSHANAVTNKYCGGGVYYRGLVEFSNICQNDCSYCGIRNNQKEVWRYTMPVEEVVEVAKWALENGI | GATANLDREQVLGVLEAVWRRGDLNLERALYSHANAVTNKYCGGGVYYRGLVEFSNICQNDCSYCGIRNNQKEVWRYTMPVEEVVEVAKWALENGI | ||
| Line 44: | Line 51: | ||
===References=== | ===References=== | ||
[1] King, P., Posewitz, M., Ghirardi, M. and Seibert, M. (2006). Functional Studies of [FeFe] Hydrogenase Maturation in an Escherichia coli Biosynthetic System. Journal of Bacteriology, 188(6), pp.2163-2172. | [1] King, P., Posewitz, M., Ghirardi, M. and Seibert, M. (2006). Functional Studies of [FeFe] Hydrogenase Maturation in an Escherichia coli Biosynthetic System. Journal of Bacteriology, 188(6), pp.2163-2172. | ||
| − | <br> | + | <br><br> |
[2] Posewitz, M., King, P., Smolinski, S., Zhang, L., Seibert, M. and Ghirardi, M. (2004). Discovery of Two Novel Radical S-Adenosylmethionine Proteins Required for the Assembly of an Active [Fe] Hydrogenase. Journal of Biological Chemistry, 279(24), pp.25711-25720. | [2] Posewitz, M., King, P., Smolinski, S., Zhang, L., Seibert, M. and Ghirardi, M. (2004). Discovery of Two Novel Radical S-Adenosylmethionine Proteins Required for the Assembly of an Active [Fe] Hydrogenase. Journal of Biological Chemistry, 279(24), pp.25711-25720. | ||
| − | <br> | + | <br><br> |
[3] Kelly, C., Pinske, C., Murphy, B., Parkin, A., Armstrong, F., Palmer, T. and Sargent, F. (2015). Integration of an [FeFe]-hydrogenase into the anaerobic metabolism of Escherichia coli. Biotechnology Reports, 8, pp.94-104. | [3] Kelly, C., Pinske, C., Murphy, B., Parkin, A., Armstrong, F., Palmer, T. and Sargent, F. (2015). Integration of an [FeFe]-hydrogenase into the anaerobic metabolism of Escherichia coli. Biotechnology Reports, 8, pp.94-104. | ||
| − | <br> | + | <br><br> |
[4] Posewitz, M., King, P., Smolinski, S., Smith, R., Ginley, A., Ghirardi, M. and Seibert, M. (2005). Identification of genes required for hydrogenase activity in Chlamydomonas reinhardtii : Figure 1. Biochm. Soc. Trans., 33(1), pp.102-104. | [4] Posewitz, M., King, P., Smolinski, S., Smith, R., Ginley, A., Ghirardi, M. and Seibert, M. (2005). Identification of genes required for hydrogenase activity in Chlamydomonas reinhardtii : Figure 1. Biochm. Soc. Trans., 33(1), pp.102-104. | ||
| − | |||
Latest revision as of 00:39, 2 November 2017
HydEF
The wrong sequence was submitted to this page in 2016. iGEM Macquarie 2017 has created a new updated part page with the correct sequence (BBa_K2300003) (https://parts.igem.org/Part:BBa_K2300003#Overview).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NotI site found at 253
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 903
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 304
Illegal BsaI.rc site found at 416
Overview
The HydEF protein contains two unique domains that are homologous to two distinct prokaryotic proteins, HydE and HydF, which are found exclusively in organisms containing [Fe] hydrogenase. This part makes up one of the parts found in an operon of the hydrogen production pathway.

Biology & Literature
hydEF (comprising of HydE and HydF) encodes proteins necessary for hydrogenase activity by functioning in H cluster biosynthesis and enzyme maturation involving S-adenosylmethionine (SAM) dependent mechanisms [1]. HydF is part of the GTPase protein family, with GTP-binding motifs present at the N terminal end and iron-sulfur cluster-binding motifs at the C terminal end [2]. Both HydE is a radical SAM (S-adenosyl methionine) enzyme [3]. King et al. stated that challenges faced during the expression of maturation genes HydEF and HydG in E. coli were that they were unstable and prone to rearrangements [1].
Both HydEF and HydG are transcribed under anaerobic conditions and are necessary for the assembly of an active HydA1 hydrogenase [2]. The importance of HydEF in hydrogenase activity has also been illustrated in the mutant hydEF-1 strain of C. reinhardtii which is unable to produce H2 due to a non-functional HydEF gene [4].
Part Verification
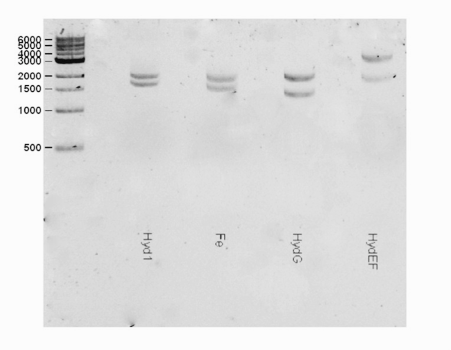
Protein information
HydEF
Mass: 121.95 kDa
Sequence:
MAHSLSAHSRQAGDRKLGAGAASSRPSCPSRRIVRVAAHASASKATPDVPVDDLPPAHARAAVAAANRRARAMASAEAAAETLGDFLGLGKGGLSP
GATANLDREQVLGVLEAVWRRGDLNLERALYSHANAVTNKYCGGGVYYRGLVEFSNICQNDCSYCGIRNNQKEVWRYTMPVEEVVEVAKWALENGI
RNIMLQGGELKTEQRLAYLEACVRAIREETTQLDLEMRARAASTTTAEAAASAQADAEAKRGEPELGVVVSLSVGELPMEQYERLFRAGARRYLIRIET
SNPDLYAALHPEPMSWHARVECLRNLKKAGYMLGTGVMVGLPGQTLHDLAGDVMFFRDIKADMIGMGPFITQPGTPATDKWTALYPNANKNSHMK
SMFDLTTAMNALVRITMGNVNISATTALQAIIPTGREIALERGANVVMPILTPTQYRESYQLYEGKPCITDTAVQCRRCLDMRLHSVGKTSAAGVWGDPA
SFLHPIVGVPVPHDLSSPALAAAASADFHEVGAGPWNPIRLERLVEVPDRYPDPDNHGRKKAGAGKGGKAHDSHDDGDHDDHHHHHGAAPAGAAA
GKGTGAAAIGGGAGASRQRVAGAAAASARLCAGARRAGRVVASPLRPAAACRGVAVKAAAAAAGEDAGAGTSGVGSNIVTSPGIASTTAHGVPRINI
GVFGVMNAGKSTLVNALAQQEACIVDSTPGTTADVKTVLLELHALGPAKLLDTAGLDEVGGLGDKKRRKALNTLKECDVAVLVVDTDTAAAAIKSGRLA
EALEWESKVMEQAHKYNVSPVLLLNVKSRGLPEAQAASMLEAVAGMLDPSKQIPRMSLDLASTPLHERSTITSAFVKEGAVRSSRYGAPLPGCLPRW
SLGRNARLLMVIPMDAETPGGRLLRPQAQVMEEAIRHWATVLSVRLDLDAARGKLGPEACEMERQRFDGVIAMMERNDGPTLVVTDSQAIDVVHPW
TLDRSSGRPLVPITTFSIAMAYQQNGGRLDPFVEGLEALETLQDGDRVLISEACNHNRITSACNDIGMVQIPNKLEAALGGKKLQIEHAFGREFPELESG
GMDGLKLAIHCGGCMIDAQKMQQRMKDLHEAGVPVTNYGVFFSWAAWPDALRRALEPWGVEPPVGTPATPAAAPATAASGV
References
[1] King, P., Posewitz, M., Ghirardi, M. and Seibert, M. (2006). Functional Studies of [FeFe] Hydrogenase Maturation in an Escherichia coli Biosynthetic System. Journal of Bacteriology, 188(6), pp.2163-2172.
[2] Posewitz, M., King, P., Smolinski, S., Zhang, L., Seibert, M. and Ghirardi, M. (2004). Discovery of Two Novel Radical S-Adenosylmethionine Proteins Required for the Assembly of an Active [Fe] Hydrogenase. Journal of Biological Chemistry, 279(24), pp.25711-25720.
[3] Kelly, C., Pinske, C., Murphy, B., Parkin, A., Armstrong, F., Palmer, T. and Sargent, F. (2015). Integration of an [FeFe]-hydrogenase into the anaerobic metabolism of Escherichia coli. Biotechnology Reports, 8, pp.94-104.
[4] Posewitz, M., King, P., Smolinski, S., Smith, R., Ginley, A., Ghirardi, M. and Seibert, M. (2005). Identification of genes required for hydrogenase activity in Chlamydomonas reinhardtii : Figure 1. Biochm. Soc. Trans., 33(1), pp.102-104.
