Difference between revisions of "Part:BBa K2276010"
| (4 intermediate revisions by 2 users not shown) | |||
| Line 16: | Line 16: | ||
===Functional Parameters=== | ===Functional Parameters=== | ||
<partinfo>BBa_K2276010 parameters</partinfo> | <partinfo>BBa_K2276010 parameters</partinfo> | ||
| − | |||
| − | |||
| − | |||
The part was designed based on the Part: BBa_I13521, while the RBS binding site is modified. The RBS is special designed for strainS whose last 9 nucleotides of the 16S rRNA are ACCTCCTTA. And it can be regard as a improvement for it has a high translation rate compared with Part: BBa_I13521.The promoter is TetR repressible promoter which is also used in the repressilator system. And the mRFP served as the reporter to help us measure the translation rate of the designed RBS. | The part was designed based on the Part: BBa_I13521, while the RBS binding site is modified. The RBS is special designed for strainS whose last 9 nucleotides of the 16S rRNA are ACCTCCTTA. And it can be regard as a improvement for it has a high translation rate compared with Part: BBa_I13521.The promoter is TetR repressible promoter which is also used in the repressilator system. And the mRFP served as the reporter to help us measure the translation rate of the designed RBS. | ||
| Line 25: | Line 22: | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | ||
| − | + | ||
| − | + | ||
| Line 35: | Line 30: | ||
<!-- --> | <!-- --> | ||
===Measurement=== | ===Measurement=== | ||
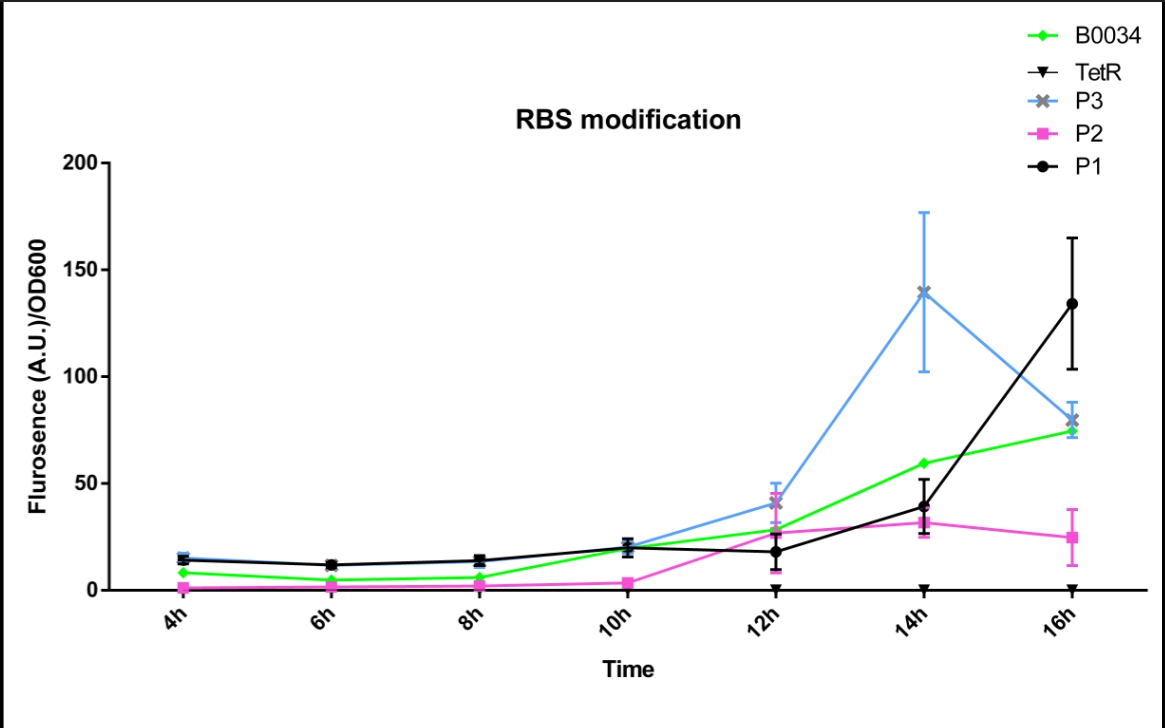
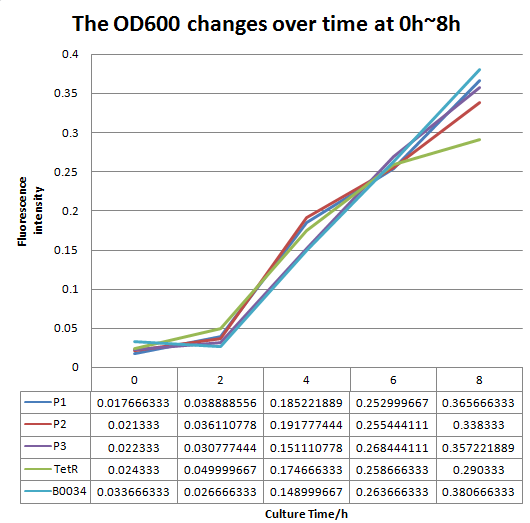
| − | + | In order to improve the expression of the original part:BBa_I13521, we predicted a synthetic RBS sequence, named P3, with much higher translation initiation rate via the RBS Calculator designed by H.M. Salis, and constructed this part BBa_K2276010 containing RBS P3. | |
| + | And then, we measured the fluorescence intensity and OD600 changes of it. As Fig 1 and Fig 2 showed,Part: the expression of BBa_K2276010 was much stronger than the original part: BBa_I13521. | ||
| + | In addition, we calculated the relative RBS strength via dividing the ratio of fluorescence intensity and OD 600 by fluorescence intensity of B0034. And this result further illustrated that this part contained a obviously stronger RBS than BBa_B0034. Thus, the function of the original part BBa_I13521 was indeed improved by us. | ||
[[File:FT3.png|500px|thumb|left|'''fig.1'''The fluorescence intensity changes over time (0h~16h). P1 represent the strain containing part: BBa_K2276007. P2 represent the strain containing part: BBa_K2276008. P3 represent the strain containing part: BBa_K2276010. TetR represent the strain that only has tetR repressible promoter without RBS and protein coding sequence. B0034 represent the strain that containing Part: BBa_I13521. | [[File:FT3.png|500px|thumb|left|'''fig.1'''The fluorescence intensity changes over time (0h~16h). P1 represent the strain containing part: BBa_K2276007. P2 represent the strain containing part: BBa_K2276008. P3 represent the strain containing part: BBa_K2276010. TetR represent the strain that only has tetR repressible promoter without RBS and protein coding sequence. B0034 represent the strain that containing Part: BBa_I13521. | ||
Latest revision as of 23:22, 1 November 2017
a composite of TetR repressible promoter、special designed RBS and the coding sequence of mRFP
The part was designed based on the Part:BBa_I13521,while the RBS binding site is modified. The RBS is special designed for strainS whose last 9 nucleotides of the 16S rRNA is ACCTCCTTA.And it can be regard as a improvement for it has a high translation rate compared with Part:BBa_I13521.The promoter is TetR repressible promoter which is also used in the repressilator system. And the mRFP served as the reporter to help us measure the translation rate of the designed RBS.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 635
Illegal AgeI site found at 747 - 1000COMPATIBLE WITH RFC[1000]
Measurement
In order to improve the expression of the original part:BBa_I13521, we predicted a synthetic RBS sequence, named P3, with much higher translation initiation rate via the RBS Calculator designed by H.M. Salis, and constructed this part BBa_K2276010 containing RBS P3. And then, we measured the fluorescence intensity and OD600 changes of it. As Fig 1 and Fig 2 showed,Part: the expression of BBa_K2276010 was much stronger than the original part: BBa_I13521. In addition, we calculated the relative RBS strength via dividing the ratio of fluorescence intensity and OD 600 by fluorescence intensity of B0034. And this result further illustrated that this part contained a obviously stronger RBS than BBa_B0034. Thus, the function of the original part BBa_I13521 was indeed improved by us.
Source
BBa_I13521
References
[1] Borujeni A E, Salis H M. Translation Initiation is Controlled by RNA Folding Kinetics via a Ribosome Drafting Mechanism[J]. Journal of the American Chemical Society, 2016, 138(22):7016.
