Difference between revisions of "Part:BBa C0060:Experience"
(→User Reviews) |
(→Applications of BBa_C0060) |
||
| Line 4: | Line 4: | ||
===Applications of BBa_C0060=== | ===Applications of BBa_C0060=== | ||
| + | The characterization of BBa_C0060 was carried out, first, by creating a composite part (BBa_K2471000) capable of expressing the biopart of interest; this was done by the addition of a T7 promoter and RBS (BBa_K525998) and a T1 terminator (BBa_B0010) to the basic part. The design was first done in silico utilizing SnapGene®, where the program indicated that the plasmid would have a length of 3,010 base pairs. After verifying the feasibility of creating this BioBrick®, the part was synthesized using the 3A assembly method. Agarose gel (1%) electrophoresis was performed to corroborate that the obtained molecular weight coincided with the expected one. | ||
===User Reviews=== | ===User Reviews=== | ||
Revision as of 20:42, 1 November 2017
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_C0060
The characterization of BBa_C0060 was carried out, first, by creating a composite part (BBa_K2471000) capable of expressing the biopart of interest; this was done by the addition of a T7 promoter and RBS (BBa_K525998) and a T1 terminator (BBa_B0010) to the basic part. The design was first done in silico utilizing SnapGene®, where the program indicated that the plasmid would have a length of 3,010 base pairs. After verifying the feasibility of creating this BioBrick®, the part was synthesized using the 3A assembly method. Agarose gel (1%) electrophoresis was performed to corroborate that the obtained molecular weight coincided with the expected one.
User Reviews
UNIQ4fae13bacd611920-partinfo-00000000-QINU
|
•••••
UNIPV-Pavia iGEM 2011 |
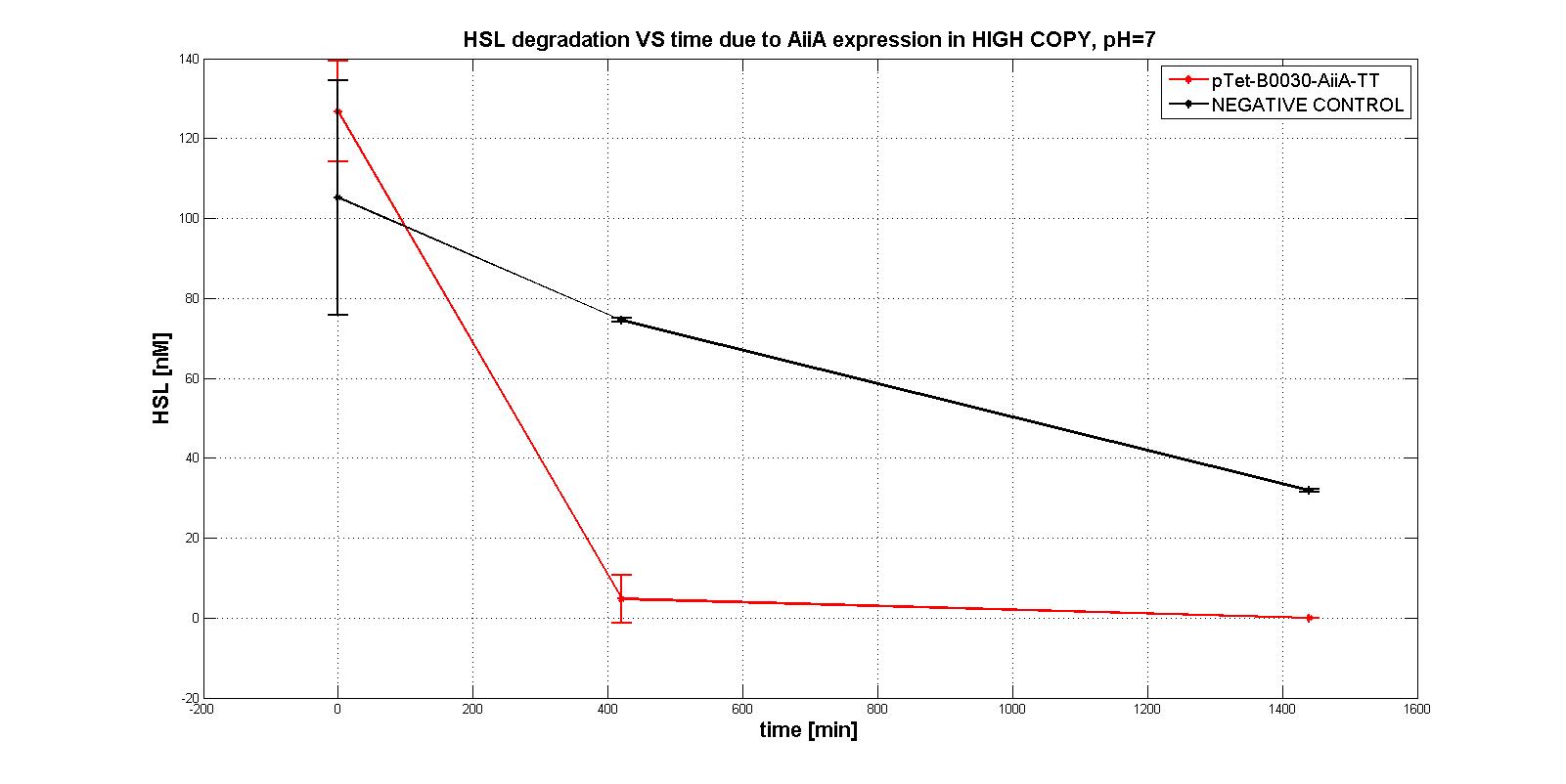
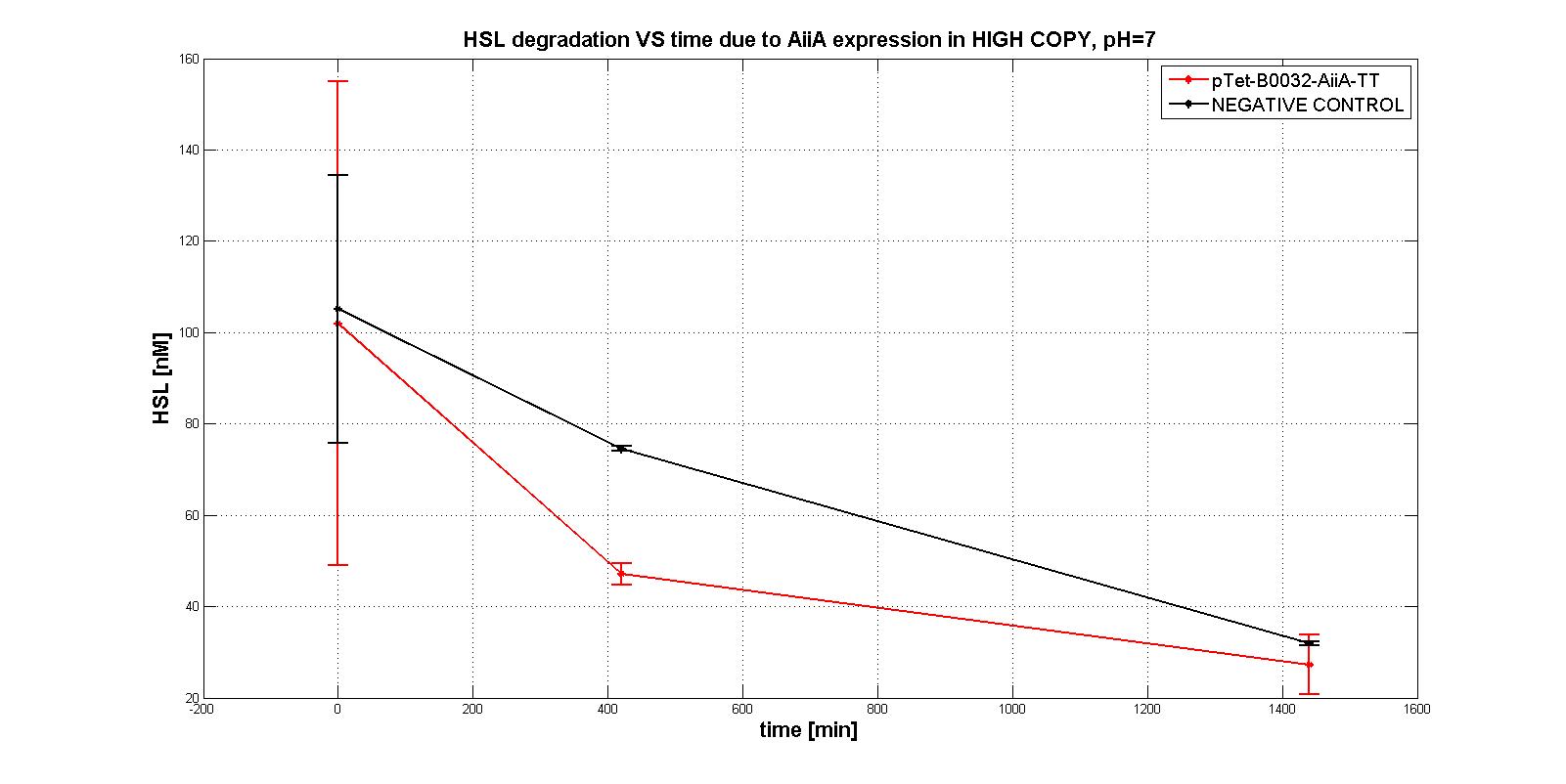
NB: unless differently specified, all tests were performed in E. coli MGZ1 in M9 supplemented medium at 37°C in low copy plasmid pSB4C5.
A system of differential equations has been derived and parameters would have been identified, studying the exponential growth phase: The four measurment parts ptet-RBSx-LuxI were quantitatively characetrized using the BBa_T9002 biosensor. Unfortunately the model parameters (kcat, kM,AiiA) were not estimated because placing the device in low copy plasmid (pSB4C5)resulted in no HSL degradation, as shown in the figures below:
In order to evaluate the RBSs efficiency, the ratio between the percentage of the degraded HSL realtive to the measurement device with the standard RBS (BBa_B0034) was computed:
|
UNIQ4fae13bacd611920-partinfo-00000003-QINU