Difference between revisions of "Part:BBa K2350021"
(→Functional Test) |
|||
| (26 intermediate revisions by one other user not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2350021 short</partinfo> | <partinfo>BBa_K2350021 short</partinfo> | ||
| − | === | + | ===Holin-Endolysin-NrtA=== |
| − | + | In this part, we combine holin (BBa_K2350019), endolysin (BBa_K2350020), and NrtA (BBa_K2350004). NrtA protein can stick to periplasmic membrane through a flexible linker to capture nitrite or nitrate in the periplasm. Therefore, we make use of NrtA in nitrogen starvation in our 2017 NYMU project. After we reach our goal, we need to kill E.coli to prevent contamination. When the lactose is added into the environment, the suicide mechanism, holin and endolyin, is induced. For more detail information, please see in BBa_2350004 for NrtA part, BBa_K2350018 for endolysin part, BBa_K2350019 for holin part, and BBa_K2350020 for endolysin-holin part. | |
===Promoter, RBS, and Terminator=== | ===Promoter, RBS, and Terminator=== | ||
| + | For more details of the connected promoter, RBS, and terminator, you can see in BBa_2350004 for NrtA part, BBa_K2350018 for endolysin part, and BBa_K2350019 for holin part. | ||
| − | |||
| − | |||
| − | + | ===Result=== | |
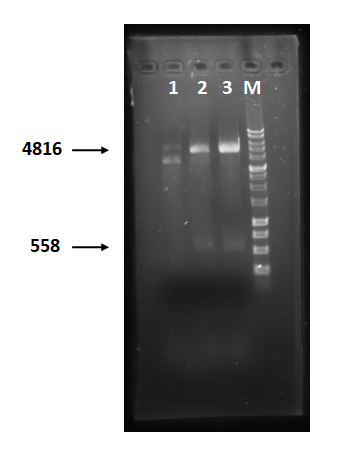
| − | + | Figure 1 is restriction enzyme check electrophoresis result of Holin-Endolysin-NrtA construct. We use XmaI to digest sample 1~3. The result shows that 2 and 3 are right. M represents 1 kb marker. | |
| − | + | ||
| − | < | + | <b>Figure 1 </b> |
| − | ===Functional | + | |
| − | < | + | [[File:Holin-Endolysin-NrtA REcheck.png|200px]] |
| − | + | ||
| + | ===Functional Test=== | ||
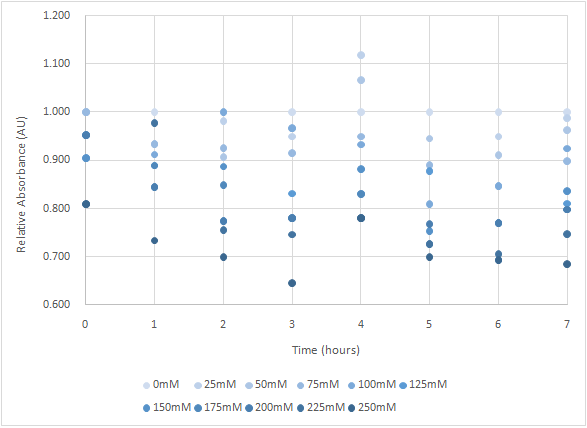
| + | As figure 2 shows, the trend of the relative absorbance is downward as the lactose is added to induce the suicide mechanism. The concentration of lactose is also positively correlated with the declining degree of relative absorbance (Experiment and graph provided by TAS_Taipei). | ||
| + | |||
| + | |||
| + | <b>Figure 2 </b> | ||
| + | |||
| + | [[File:T--NYMU-Taipei--TAS-FunctionalTest.png|700px]] | ||
| + | |||
| + | |||
| + | <partinfo>BBa_K2350021 SequenceAndFeatures</partinfo> | ||
Latest revision as of 09:36, 1 November 2017
R0010-B0034-Holin-B0010-B0012-J23106-B0034-Endolysin-B0010-B0012-J23118-B0034-NrtA-B0015
Holin-Endolysin-NrtA
In this part, we combine holin (BBa_K2350019), endolysin (BBa_K2350020), and NrtA (BBa_K2350004). NrtA protein can stick to periplasmic membrane through a flexible linker to capture nitrite or nitrate in the periplasm. Therefore, we make use of NrtA in nitrogen starvation in our 2017 NYMU project. After we reach our goal, we need to kill E.coli to prevent contamination. When the lactose is added into the environment, the suicide mechanism, holin and endolyin, is induced. For more detail information, please see in BBa_2350004 for NrtA part, BBa_K2350018 for endolysin part, BBa_K2350019 for holin part, and BBa_K2350020 for endolysin-holin part.
Promoter, RBS, and Terminator
For more details of the connected promoter, RBS, and terminator, you can see in BBa_2350004 for NrtA part, BBa_K2350018 for endolysin part, and BBa_K2350019 for holin part.
Result
Figure 1 is restriction enzyme check electrophoresis result of Holin-Endolysin-NrtA construct. We use XmaI to digest sample 1~3. The result shows that 2 and 3 are right. M represents 1 kb marker.
Figure 1
Functional Test
As figure 2 shows, the trend of the relative absorbance is downward as the lactose is added to induce the suicide mechanism. The concentration of lactose is also positively correlated with the declining degree of relative absorbance (Experiment and graph provided by TAS_Taipei).
Figure 2
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1035
Illegal NheI site found at 1058
Illegal NheI site found at 1757
Illegal NheI site found at 1780 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 2990
Illegal BamHI site found at 2360 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 1362
Illegal AgeI site found at 1432 - 1000COMPATIBLE WITH RFC[1000]