Difference between revisions of "Part:BBa K1893013"
Georgedashen (Talk | contribs) (→New design) |
|||
| (37 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K1893013 short</partinfo> | <partinfo>BBa_K1893013 short</partinfo> | ||
| − | This is the STAR construct with its own transcription terminator t500. When transcribed, it produces the STAR-antisense RNA | + | This is the STAR (Small Transcription Activating RNA) construct with its own transcription terminator t500 [Chappell et al., 2015]. STAR is a small bacterial RNA (68 nucleotide-long) that can be used to directly activate transcription. It is a novel RNA-logic technology that allows for rapid and robust regulation. When transcribed, it produces the STAR-antisense RNA, which can bind to the sense STAR-target terminator attenuator sequence. Thus, only in the presence of STAR-antisense, transcription and expression of coding sequences downstream to the STAR-target is activated. |
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | < | + | For designing a small transcriptional activating RNA through an anti-terminator mechanism, Chappell et. al. investigated transcriptional terminators from different plasmids and of different lengths and sequences. Of these, the sense pAD1 plasmid attenuator sequence no. 5 (pAD1.S5) and its associated STAR-antisense sequence no. 5 (pAD1.A5) pair gave the highest (94-fold) activation of the reporter gene, SFGFP [Chappell et al., 2015]. Thus, we used the pAD1.S5 sequence as our STAR target-encoding DNA and the pAD1.A5 sequence as the STAR-antisense-encoding DNA. |
| + | |||
| + | STAR binds a sense target RNA sequence, the pAD1 plasmid attenuator sequence, which is fused upstream of the gene of interest. In the absence of STAR, the pAD1 plasmid attenuator sequence forms an intrinsic hairpin structure, preventing RNA polymerase from transcribing the gene and thus acting as a terminator of transcription. However, when STAR is produced, it binds to the target sequence and interferes with the hairpin formation, allowing transcription of the downstream gene. | ||
| + | |||
| + | Imperial iGEM 2016 chose this as its best basic part as this is a completely new method of gene regulation in the BioBrick Registry, it offers significant advantages over protein-based regulatory mechanisms and it makes RNA-only circuits possible. | ||
| + | |||
| + | <b>Advantages of using STAR over traditional protein-based regulatory systems:</b> <br> | ||
| + | → <b>Low metabolic burden:</b> the RNA is functional after transcription, avoiding the resource-intensive translation steps. <br> | ||
| + | → <b>Portability:</b> trans-acting RNA based regulation is found throughout the bacterial kingdom (as well as other kingdoms) and is not host-specific. <br> | ||
| + | → <b>Tight Regulation:</b> In the absence of STAR there is very low baseline expression of the downstream gene, while in the presence of STAR the expression is activated 21-fold. <br> | ||
| + | → <b>Ease of design:</b> Watson-Crick base pairing avoids the need for structural protein-protein interactions, which are difficult to predict and design. <br> | ||
| + | → <b>Reduced lag in circuits:</b> Using RNAs increases speed of signal propagation and since its degradation is rapid, it gives short-time responses. <br> | ||
| + | |||
| + | |||
| + | ===Characterisation=== | ||
| + | |||
| + | We generated a two-plasmid system for characterisation experiments. The first plasmid contains the STAR sequence downstream of a constitutive Anderson promoter and followed by the t500 transcriptional terminator on a high-copy plasmid. The second plasmid is a reporter plasmid that contains the superfolder GFP (SFGFP) gene with a ribosome binding site immediately downstream of the STAR-target (pAD1 plasmid attenuator) sequence. The SFGFP coding sequence is under the control of a constitutive Anderson promoter and also has its own TrrnB terminator. | ||
| + | |||
| + | We characterised STAR activity at 30 and 37 degrees Celsius by using a plate reader to record the fluorescence of SFGFP over 200 minutes in cells with both plasmids, as well as in cells with just the reporter plasmid to determine to what degree STAR was able to activate SFGFP transcription. | ||
| + | |||
| + | Initially, we characterised the STAR system in terms of fold activation of GFP expression from the reporter plasmid in the absence and presence of STAR molecules. For this experiment, Top 10 E. coli cell lines were co-transformed with either the reporter plasmid and a plasmid with the J23119 promoter (no STAR) or with the reporter plasmid and the J23119-STAR plasmid. Cell cultures of these two cell lines were grown in microplates and well fluorescence was monitored over time using a microplate reader. The fluorescence signal from each well was normalised by dividing with the O.D. 600 value of that well. This gave the normalised fluorescence value for that cell line (FI/Abs). To account for cell autofluorescence, DH10B cells (similar to Top10) were used to determine background normalised fluorescence value. | ||
| + | |||
| + | Next, an experiment was carried out to investigate STAR system functionality at different temperatures. RNA elements functionality can be strongly depended on temperature and thermodynamics. In the case that we needed to use the STAR system under different circumstances (co-culture with B. subtilis that grows better at 30 degrees Celcius) we characterised the STAR system at 30 degrees Celsius cell culture condition. Cell cultures of the previously mentioned cell lines were grown for 5 hours in exponential phase. Then, the cell cultures were normalised at O.D. 600 = 0.4 and the fluorescent signal for each condition was recorded. | ||
| + | |||
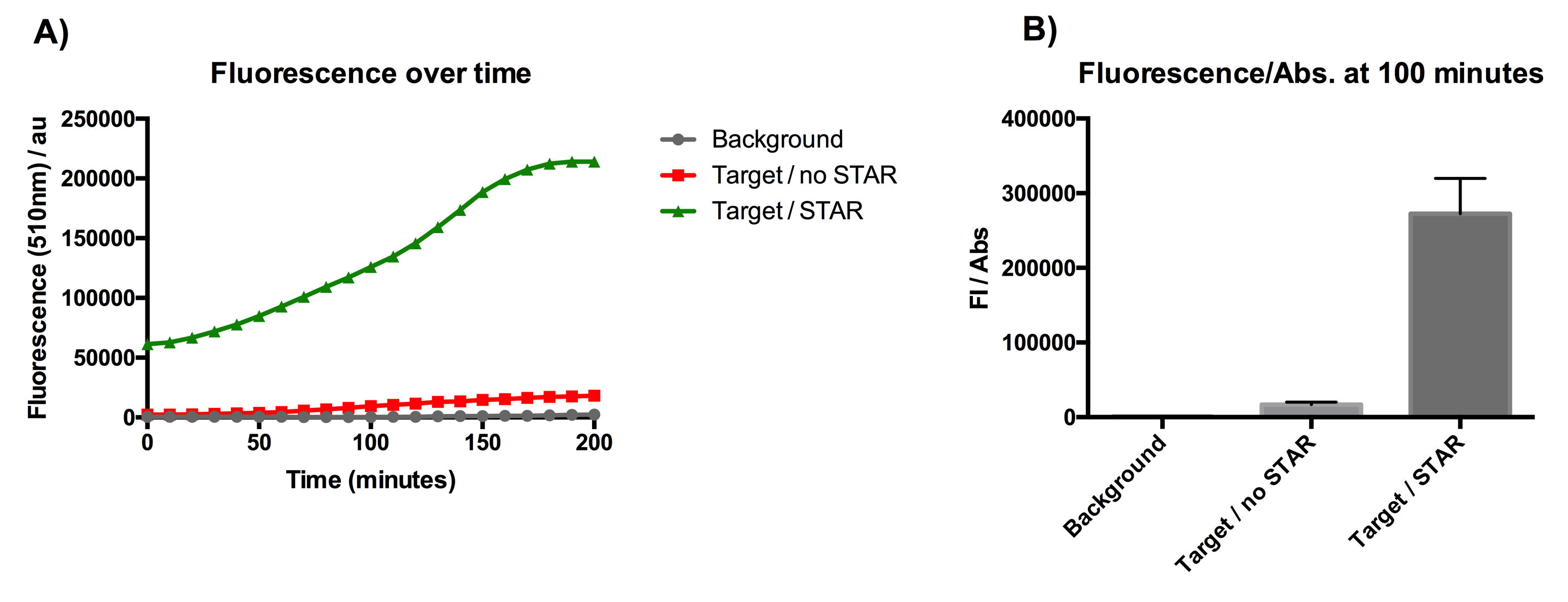
| + | [[File:IC16_STAR1stgraph.png|700px|center|]] | ||
| + | Figure 1: Characterisation of STAR system in TOP10 E. coli cells. (A) Normalised fluorescence monitored over time for cell lines incorporating the STAR system in the absence or presence of transcribed STAR molecules (B) Normalised endpoint fluorescence (100 minutes) for cell lines in the absence or presence of STAR molecules. We used the two-plasmid system described in the Experimental Design for characterisation experiments involving STAR. For the absence of STAR condition, the plasmid did not include STAR sequence but just the J23119 promoter. Normalised fluorescence was calculated by dividing fluorescent signal by the O.D.600 value of the culture. Background was determined by the use of DH10B cells with no plasmid transformed. Error bars represent standard deviation from 3 technical repeats. | ||
| + | |||
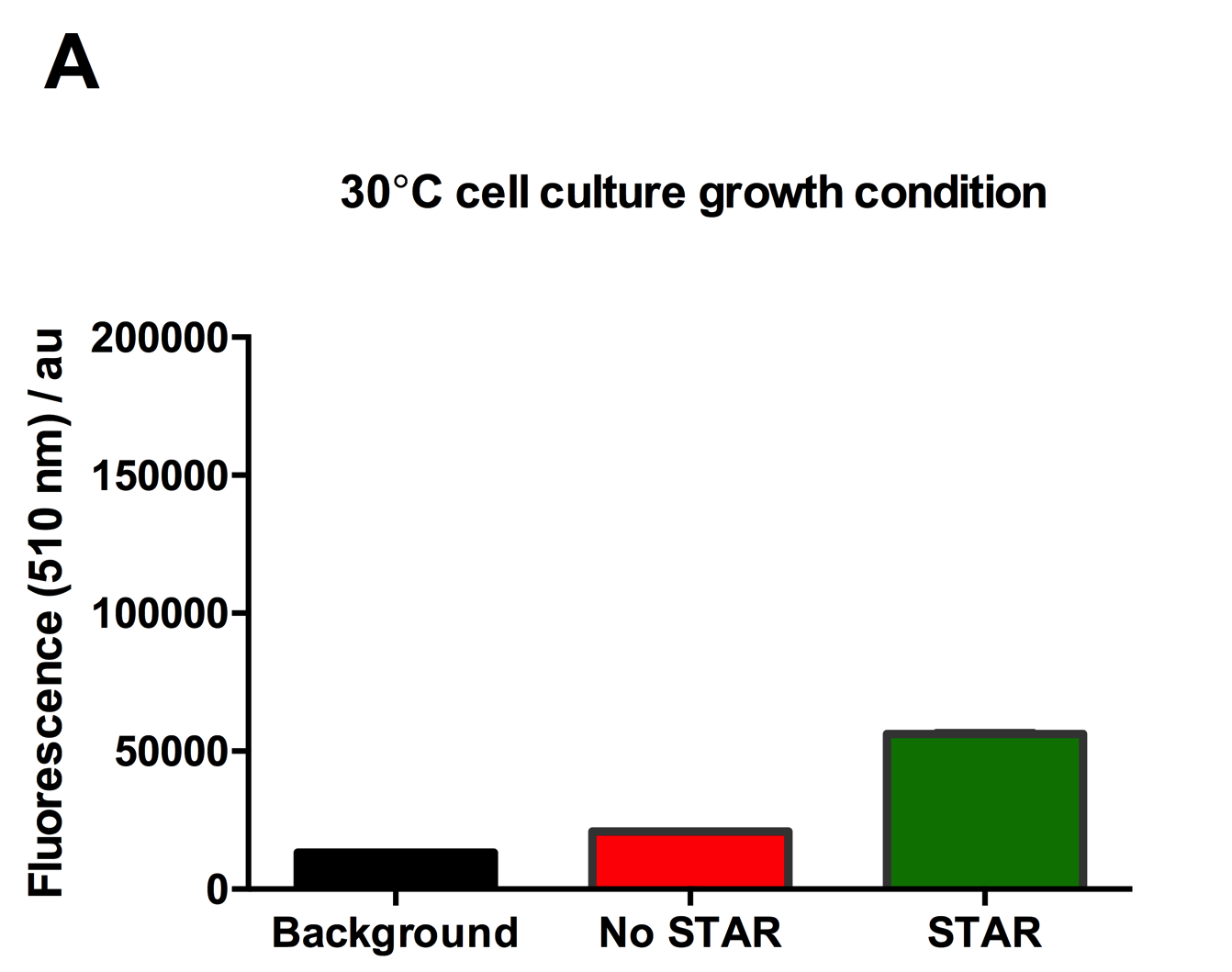
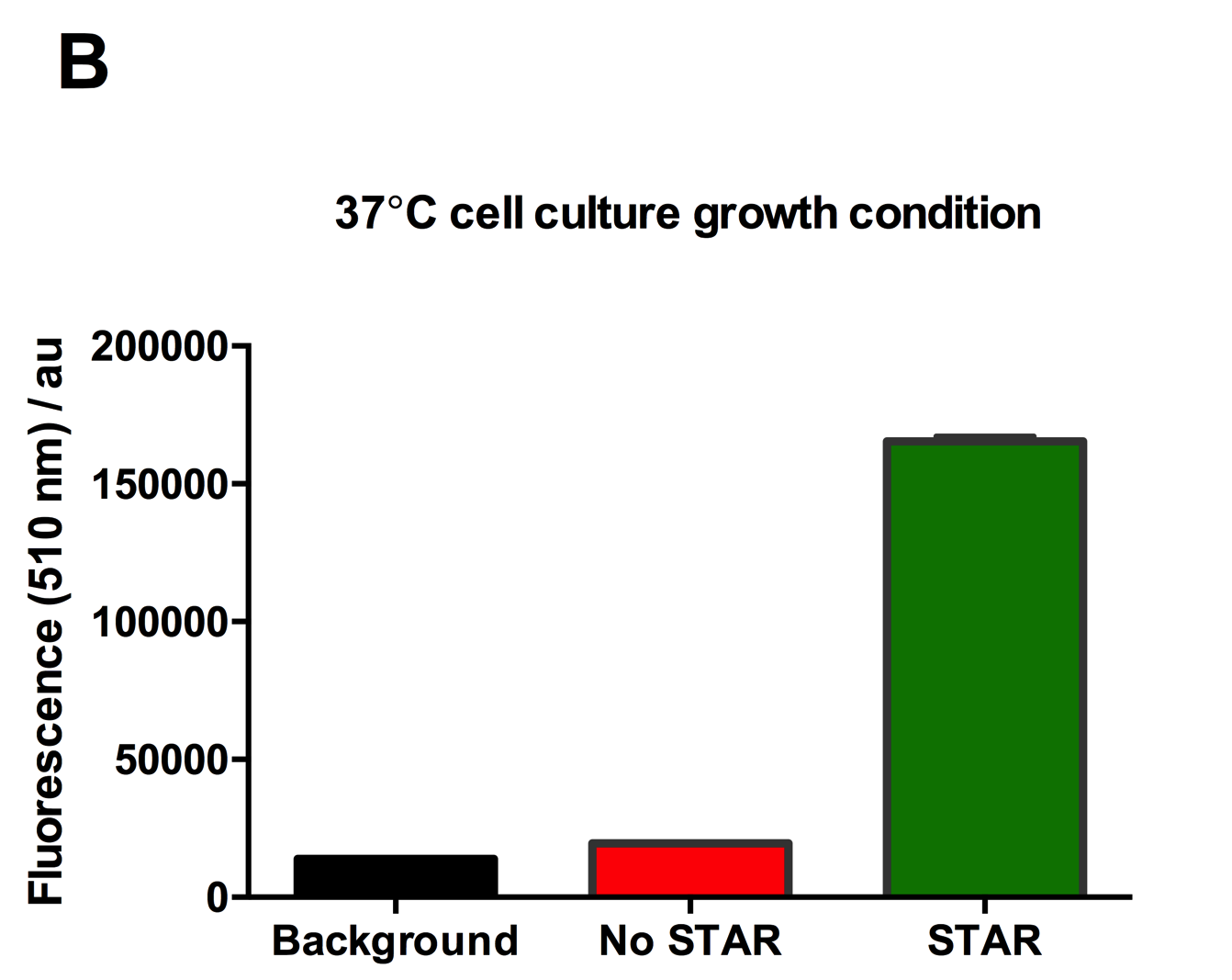
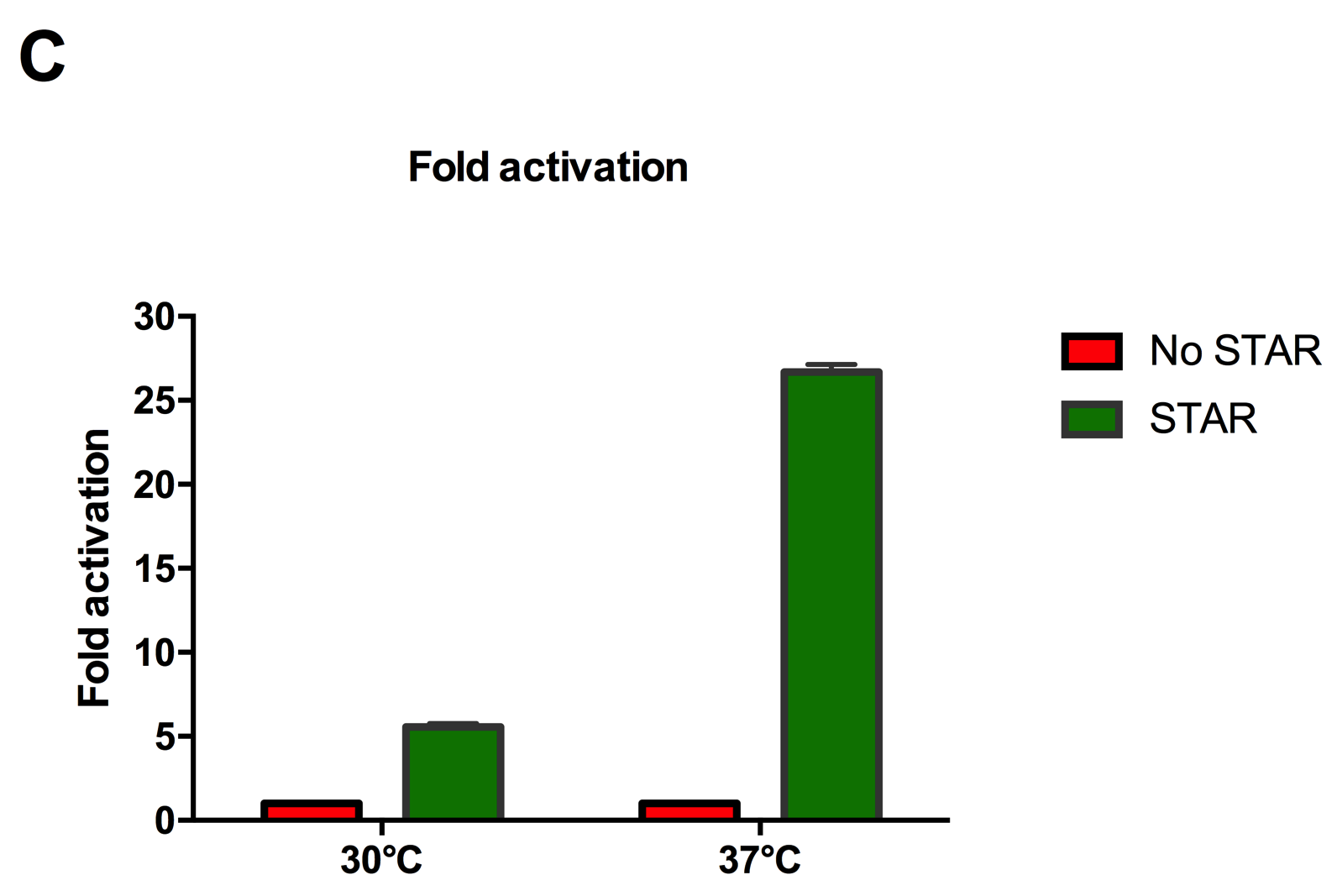
| + | [[File:IC16_STAR2Agraph.png|300px|]][[File:IC16_STAR2B.png|300px|]][[File:IC16_STAR2C.png|345px|]] | ||
| + | |||
| + | Figure 2: Characterisation of STAR system in TOP10 E. coli cells at different temperatures. (A) Cell culture fluorescence at 30°C (B) Cell culture fluorescence assay at 37°C. (C) Fold activation SFGFP expression in presence of STAR. We used the two-plasmid system described in the Experimental Design for characterisation experiments involving STAR. For the absence of STAR condition, the plasmid did not include STAR sequence but just the J23119 promoter. The autofluorescence background control used is E. coli Top 10 cells with no reporter plasmid. Error bars represent standard deviation from 3 technical repeats. | ||
| + | |||
| + | |||
| + | ===Analysis=== | ||
| + | |||
| + | The results displayed in Figure 1A show that the expression of SFGFP is greatly increased in the presence of STAR over the course of 200 minutes of culturing. The graph from Figure 1B represents the normalised fluorescence once the growth data (optical density) has been taken into account. The data from this graph shows that SFGFP expression is increased more than 17-fold at 100 minutes of culturing, when STAR is present. This indicates that STAR is successful in preventing the pAD1 plasmid attenuator from interfering with SFGFP transcription. The data suggests that STAR is a promising tool for the regulation of the growth repressing gene in our circuit. | ||
| + | |||
| + | Experimental results showed that STAR system activated transcription at various levels at different temperatures. Figure 2A shows that SFGFP expression at 30°C increases significantly in the presence of STAR molecules as determined by the cell culture fluorescence. Figure 2B shows that SFGFP expression at 37°C increases significantly in the presence of STAR molecules as determined by the cell culture fluorescence, in agreement with our previous results. Figure 2C shows that the activation SFGFP expression attainable by the use of the STAR system was much greater at 37°C (26.7 fold) rather than at 30°C (5.5 fold). The results showcase important considerations that need to be taken into account when the STAR system is used under different culture conditions, and can be incorporated into the models of the system to inform design. | ||
| + | |||
| + | |||
| + | ===New design=== | ||
| + | |||
| + | '''iGEM17_SJTU_BioX_Shanghai:''' | ||
| + | |||
| + | '''STAR3 '''<partinfo>BBa_K2285020</partinfo> | ||
| + | |||
| + | This summer, we designed another STAR system to achieve multiple factors detection. To make these systems more convenient, we constructed both Target and STAR(called Antisense in our project) on the same plasmid. In order to know whether the characteristics change, we conducted the same experiment again but in our dual-expression plasmid pCDF-Duet1 based on the characterization above. To distinguish two systems, we call our new STAR system STAR3 and the STAR system used by Imperial College STAR1. | ||
| + | |||
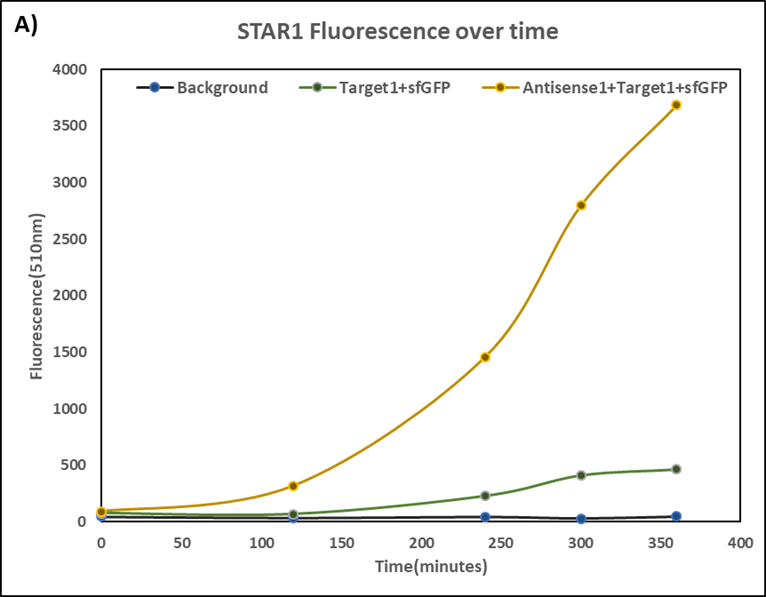
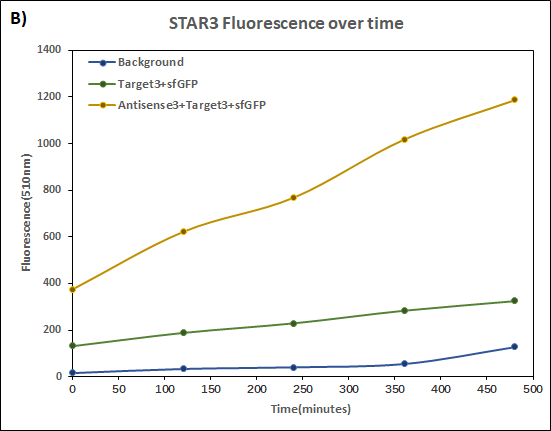
| + | [[Image:T1A1.jpg|420px]][[Image:T3A3.jpg|418px]] | ||
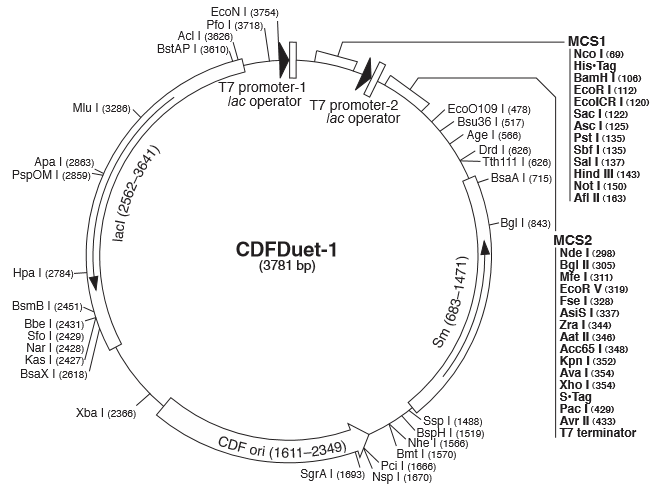
| + | [[Image:pCDF.JPG|350px]] | ||
| + | |||
| + | '''Figure 1(A) (B) : '''Characterization of STARs system in DH5α strain with pCDF as vector. Nomalized fluorescence over time for cell growth incorporating Star1 system in the absence or presence of transcribed Antisense Molecules. We used the dual-expression plasmid pCDF-Duet1. For the absence of Antisense1, the plasmid just includes Target1 in the MCS1. For the presence of Antisense1, Antisense1 is inserted into MCS2. For control(background), only pCDF backbone is transformed into DH5α.''' Figure2: '''Map of plasmid pCDF. | ||
| + | |||
| + | |||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K1893013 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1893013 SequenceAndFeatures</partinfo> | ||
Latest revision as of 08:40, 1 November 2017
Small transcription activating RNA (STAR)
This is the STAR (Small Transcription Activating RNA) construct with its own transcription terminator t500 [Chappell et al., 2015]. STAR is a small bacterial RNA (68 nucleotide-long) that can be used to directly activate transcription. It is a novel RNA-logic technology that allows for rapid and robust regulation. When transcribed, it produces the STAR-antisense RNA, which can bind to the sense STAR-target terminator attenuator sequence. Thus, only in the presence of STAR-antisense, transcription and expression of coding sequences downstream to the STAR-target is activated.
Usage and Biology
For designing a small transcriptional activating RNA through an anti-terminator mechanism, Chappell et. al. investigated transcriptional terminators from different plasmids and of different lengths and sequences. Of these, the sense pAD1 plasmid attenuator sequence no. 5 (pAD1.S5) and its associated STAR-antisense sequence no. 5 (pAD1.A5) pair gave the highest (94-fold) activation of the reporter gene, SFGFP [Chappell et al., 2015]. Thus, we used the pAD1.S5 sequence as our STAR target-encoding DNA and the pAD1.A5 sequence as the STAR-antisense-encoding DNA.
STAR binds a sense target RNA sequence, the pAD1 plasmid attenuator sequence, which is fused upstream of the gene of interest. In the absence of STAR, the pAD1 plasmid attenuator sequence forms an intrinsic hairpin structure, preventing RNA polymerase from transcribing the gene and thus acting as a terminator of transcription. However, when STAR is produced, it binds to the target sequence and interferes with the hairpin formation, allowing transcription of the downstream gene.
Imperial iGEM 2016 chose this as its best basic part as this is a completely new method of gene regulation in the BioBrick Registry, it offers significant advantages over protein-based regulatory mechanisms and it makes RNA-only circuits possible.
Advantages of using STAR over traditional protein-based regulatory systems:
→ Low metabolic burden: the RNA is functional after transcription, avoiding the resource-intensive translation steps.
→ Portability: trans-acting RNA based regulation is found throughout the bacterial kingdom (as well as other kingdoms) and is not host-specific.
→ Tight Regulation: In the absence of STAR there is very low baseline expression of the downstream gene, while in the presence of STAR the expression is activated 21-fold.
→ Ease of design: Watson-Crick base pairing avoids the need for structural protein-protein interactions, which are difficult to predict and design.
→ Reduced lag in circuits: Using RNAs increases speed of signal propagation and since its degradation is rapid, it gives short-time responses.
Characterisation
We generated a two-plasmid system for characterisation experiments. The first plasmid contains the STAR sequence downstream of a constitutive Anderson promoter and followed by the t500 transcriptional terminator on a high-copy plasmid. The second plasmid is a reporter plasmid that contains the superfolder GFP (SFGFP) gene with a ribosome binding site immediately downstream of the STAR-target (pAD1 plasmid attenuator) sequence. The SFGFP coding sequence is under the control of a constitutive Anderson promoter and also has its own TrrnB terminator.
We characterised STAR activity at 30 and 37 degrees Celsius by using a plate reader to record the fluorescence of SFGFP over 200 minutes in cells with both plasmids, as well as in cells with just the reporter plasmid to determine to what degree STAR was able to activate SFGFP transcription.
Initially, we characterised the STAR system in terms of fold activation of GFP expression from the reporter plasmid in the absence and presence of STAR molecules. For this experiment, Top 10 E. coli cell lines were co-transformed with either the reporter plasmid and a plasmid with the J23119 promoter (no STAR) or with the reporter plasmid and the J23119-STAR plasmid. Cell cultures of these two cell lines were grown in microplates and well fluorescence was monitored over time using a microplate reader. The fluorescence signal from each well was normalised by dividing with the O.D. 600 value of that well. This gave the normalised fluorescence value for that cell line (FI/Abs). To account for cell autofluorescence, DH10B cells (similar to Top10) were used to determine background normalised fluorescence value.
Next, an experiment was carried out to investigate STAR system functionality at different temperatures. RNA elements functionality can be strongly depended on temperature and thermodynamics. In the case that we needed to use the STAR system under different circumstances (co-culture with B. subtilis that grows better at 30 degrees Celcius) we characterised the STAR system at 30 degrees Celsius cell culture condition. Cell cultures of the previously mentioned cell lines were grown for 5 hours in exponential phase. Then, the cell cultures were normalised at O.D. 600 = 0.4 and the fluorescent signal for each condition was recorded.
Figure 1: Characterisation of STAR system in TOP10 E. coli cells. (A) Normalised fluorescence monitored over time for cell lines incorporating the STAR system in the absence or presence of transcribed STAR molecules (B) Normalised endpoint fluorescence (100 minutes) for cell lines in the absence or presence of STAR molecules. We used the two-plasmid system described in the Experimental Design for characterisation experiments involving STAR. For the absence of STAR condition, the plasmid did not include STAR sequence but just the J23119 promoter. Normalised fluorescence was calculated by dividing fluorescent signal by the O.D.600 value of the culture. Background was determined by the use of DH10B cells with no plasmid transformed. Error bars represent standard deviation from 3 technical repeats.
Figure 2: Characterisation of STAR system in TOP10 E. coli cells at different temperatures. (A) Cell culture fluorescence at 30°C (B) Cell culture fluorescence assay at 37°C. (C) Fold activation SFGFP expression in presence of STAR. We used the two-plasmid system described in the Experimental Design for characterisation experiments involving STAR. For the absence of STAR condition, the plasmid did not include STAR sequence but just the J23119 promoter. The autofluorescence background control used is E. coli Top 10 cells with no reporter plasmid. Error bars represent standard deviation from 3 technical repeats.
Analysis
The results displayed in Figure 1A show that the expression of SFGFP is greatly increased in the presence of STAR over the course of 200 minutes of culturing. The graph from Figure 1B represents the normalised fluorescence once the growth data (optical density) has been taken into account. The data from this graph shows that SFGFP expression is increased more than 17-fold at 100 minutes of culturing, when STAR is present. This indicates that STAR is successful in preventing the pAD1 plasmid attenuator from interfering with SFGFP transcription. The data suggests that STAR is a promising tool for the regulation of the growth repressing gene in our circuit.
Experimental results showed that STAR system activated transcription at various levels at different temperatures. Figure 2A shows that SFGFP expression at 30°C increases significantly in the presence of STAR molecules as determined by the cell culture fluorescence. Figure 2B shows that SFGFP expression at 37°C increases significantly in the presence of STAR molecules as determined by the cell culture fluorescence, in agreement with our previous results. Figure 2C shows that the activation SFGFP expression attainable by the use of the STAR system was much greater at 37°C (26.7 fold) rather than at 30°C (5.5 fold). The results showcase important considerations that need to be taken into account when the STAR system is used under different culture conditions, and can be incorporated into the models of the system to inform design.
New design
iGEM17_SJTU_BioX_Shanghai:
STAR3 BBa_K2285020
This summer, we designed another STAR system to achieve multiple factors detection. To make these systems more convenient, we constructed both Target and STAR(called Antisense in our project) on the same plasmid. In order to know whether the characteristics change, we conducted the same experiment again but in our dual-expression plasmid pCDF-Duet1 based on the characterization above. To distinguish two systems, we call our new STAR system STAR3 and the STAR system used by Imperial College STAR1.
Figure 1(A) (B) : Characterization of STARs system in DH5α strain with pCDF as vector. Nomalized fluorescence over time for cell growth incorporating Star1 system in the absence or presence of transcribed Antisense Molecules. We used the dual-expression plasmid pCDF-Duet1. For the absence of Antisense1, the plasmid just includes Target1 in the MCS1. For the presence of Antisense1, Antisense1 is inserted into MCS2. For control(background), only pCDF backbone is transformed into DH5α. Figure2: Map of plasmid pCDF.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]