Difference between revisions of "Part:BBa K2448023"
| Line 13: | Line 13: | ||
Apart from its fast building design, the UBC has been engineered to be used in high throughput procedures such as screening. Indeed, we originally designed it for a new enzyme engineering process for D-psicose production, using a psicose biosensor to identify the enzyme mutants showing the best activity. In addition, by its design, the UBC allows fast cloning: depending on what is to be inserted into the UBC and one’s mastery of the Golden Gate Assembly, a functional biosensor can be obtained in less than a week. | Apart from its fast building design, the UBC has been engineered to be used in high throughput procedures such as screening. Indeed, we originally designed it for a new enzyme engineering process for D-psicose production, using a psicose biosensor to identify the enzyme mutants showing the best activity. In addition, by its design, the UBC allows fast cloning: depending on what is to be inserted into the UBC and one’s mastery of the Golden Gate Assembly, a functional biosensor can be obtained in less than a week. | ||
| − | For more information | + | For more information visit http://2017.igem.org/Team:Evry_Paris-Saclay |
| Line 27: | Line 27: | ||
*Strong RBSs ([[Part:BBa_B0034|BBa_BBa_B0034]]) and efficient synthetic terminators ([[Part:BBa_B0015|BBa_B0015]], [[Part:BBa_K2448018|BBa_K2448018]]). | *Strong RBSs ([[Part:BBa_B0034|BBa_BBa_B0034]]) and efficient synthetic terminators ([[Part:BBa_B0015|BBa_B0015]], [[Part:BBa_K2448018|BBa_K2448018]]). | ||
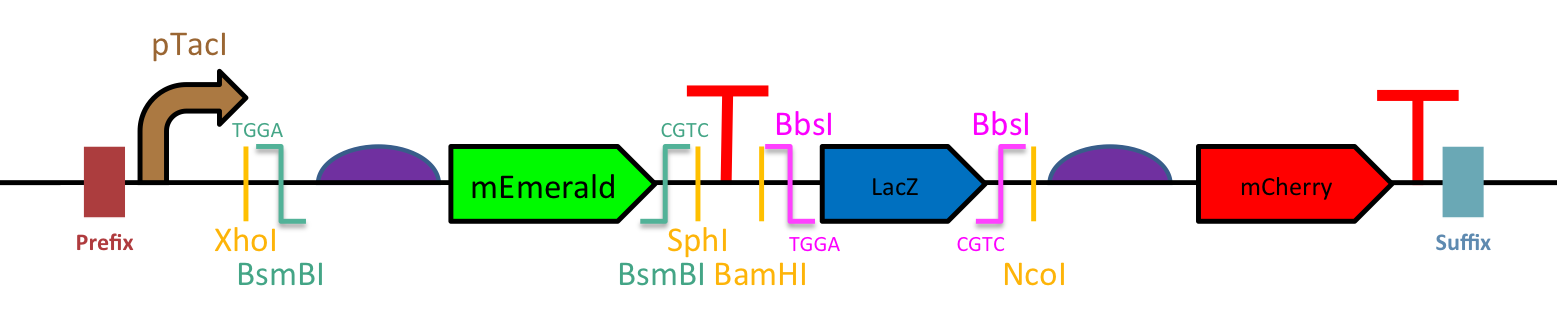
| − | [[File: T--Evry_Paris-Saclay--UBC_structure.png]] | + | [[File: T--Evry_Paris-Saclay--UBC_structure.png|600px]] |
Figure 1 : Schematic design of the UBC. | Figure 1 : Schematic design of the UBC. | ||
| + | |||
| + | For more information visit http://2017.igem.org/Team:Evry_Paris-Saclay/Composite_Part | ||
====Usage==== | ====Usage==== | ||
| Line 35: | Line 37: | ||
The Golden Gate Assembly building process combined with the presence of insertion markers naturally mark the UBC for high throughput screening experiments. It enables the characterization of many parts at once, coupled with a Fluorescence plate reader or Flow Cytometer. Indeed, insertion of various promoters or transcription factors in parallel could not only help in screening banks of promoters, RBS or transcription factors by measuring the fluorescence changes under various conditions, but also banks of enzyme mutants. By comparing the fluorescence, using the same biosensor, the more signal you observe, the more compound of interest there would be. | The Golden Gate Assembly building process combined with the presence of insertion markers naturally mark the UBC for high throughput screening experiments. It enables the characterization of many parts at once, coupled with a Fluorescence plate reader or Flow Cytometer. Indeed, insertion of various promoters or transcription factors in parallel could not only help in screening banks of promoters, RBS or transcription factors by measuring the fluorescence changes under various conditions, but also banks of enzyme mutants. By comparing the fluorescence, using the same biosensor, the more signal you observe, the more compound of interest there would be. | ||
| − | For more information | + | For more information visit http://2017.igem.org/Team:Evry_Paris-Saclay |
Revision as of 21:26, 30 October 2017
Universal Biosensing Chassis (UBC)
Universal Biosensing Chassis (UBC)
Usage and Biology
The Universal Biosensing Chassis (UBC) is the keystone of our project. It aims to provide an answer to the lack of rapid and reliable building methods for transcription-factor based biosensors. Biosensors rely on a basic theoretical principle: a certain concentration of a molecule of interest induces a proportional production of a fluorescent compound. Transcription-factor based biosensors allow the precise and cheap detection or quantification of various chemical compounds.
Using our composite biobrick, one will only need a suitable transcription factor, able to bind to the molecule of interest, and its related promoter. However sometimes, there is no transcription factor in the databases that matches what we are looking for. Here we can use indirect sensing or Sensing-Enabling Metabolic Pathways. Essentially, tools like [http://sensipath.micalis.fr Sensipath] search and design the enzymatic reactions necessary to transform your non-detectable molecule in a detectable one, therefore expanding the repertoire of molecules available to sensing.
Apart from its fast building design, the UBC has been engineered to be used in high throughput procedures such as screening. Indeed, we originally designed it for a new enzyme engineering process for D-psicose production, using a psicose biosensor to identify the enzyme mutants showing the best activity. In addition, by its design, the UBC allows fast cloning: depending on what is to be inserted into the UBC and one’s mastery of the Golden Gate Assembly, a functional biosensor can be obtained in less than a week.
For more information visit http://2017.igem.org/Team:Evry_Paris-Saclay
Features
The highly modular architecture of the UBC (Figure 1) allows rapid engineering of your biosensor:
- Standardized Fusion sites for Golden Gate Assembly: The UBC has been design to facilitate the DNA insertion hence improving speed and ease of construction. You will only need transcription factors and promoters flanked by BsmBI or BbsI, respectively, with appropriate cutting sites for these type IIS restriction enzymes (5’-TGGA and GCAG-3’).
- Many restriction sites: If Golden Gate assembly isn’t convenient for you, we have included various restriction sites to allow the insertion of promoters and transcription factors (with the RBS) using traditional digestion-ligation protocol.
- Insertion markers: In order to enable quick and easy identification of the right clones, we put two different reporter genes : mEmerald (BBa_K2448001) for the transcription factors and LacZ-alpha (BBa_K2448003) for the promoters. If markers are an issue for you, a shorter version of the UBC also exists in the registry (BBa_K2448024).
- An inducible promoter to control the transcription factor expression: pTacI (BBa_K864400). This well-known promoter is IPTG inducible and remains very strong. To better regulate its expression, and consequently the production of the transcription factor, it could be worthwhile to use our pSB1C3 LacIq (BBa_K2448038) as vector.
- An efficient reporter: mCherry (BBa_K2448004). This fluorescent monomeric protein widely used in biotechnology is derived from the RFP. Its rapid maturation, low brightness as well as its improved photostability and resistance to bleaching makes it the perfect reporter for biosensors for precise measurements. Moreover, unlike GFP like proteins, there is no cell auto-fluorescence effect at its excitation wavelength.
- Strong RBSs (BBa_BBa_B0034) and efficient synthetic terminators (BBa_B0015, BBa_K2448018).
Figure 1 : Schematic design of the UBC.
For more information visit http://2017.igem.org/Team:Evry_Paris-Saclay/Composite_Part
Usage
The Golden Gate Assembly building process combined with the presence of insertion markers naturally mark the UBC for high throughput screening experiments. It enables the characterization of many parts at once, coupled with a Fluorescence plate reader or Flow Cytometer. Indeed, insertion of various promoters or transcription factors in parallel could not only help in screening banks of promoters, RBS or transcription factors by measuring the fluorescence changes under various conditions, but also banks of enzyme mutants. By comparing the fluorescence, using the same biosensor, the more signal you observe, the more compound of interest there would be.
For more information visit http://2017.igem.org/Team:Evry_Paris-Saclay
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1033
Illegal NheI site found at 1056 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1005
Illegal XhoI site found at 62 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 128
Illegal AgeI site found at 251
Illegal AgeI site found at 2154
Illegal AgeI site found at 2266 - 1000COMPATIBLE WITH RFC[1000]