Difference between revisions of "Part:BBa K2407301"
| (23 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2407301 short</partinfo> | <partinfo>BBa_K2407301 short</partinfo> | ||
| − | This is the part regulatory region from the URA3 gene coding for OMP decarboxylase, an essential protein in the uracil synthesis pathway in S. cerevisiae budding yeast. | + | This is the part regulatory region from the URA3 gene coding for OMP decarboxylase, an essential protein in the uracil synthesis pathway in S. cerevisiae budding yeast. It is widely used as a nutrition tag in Saccharomyces cerevisiae. |
| + | |||
| + | <h4>Description</h4> | ||
| + | <p><b><i>URA3</i></b>, a gene on <i>chromosome V</i> in <i>Saccharomvces cerevisiae</i>, is widely used in researches concerning yeasts as a <i>“marker gene”</i> (systematic name <b><i>YEL021W. URA3</i></b>) and used as a label for chromosomes orplasmids. <b><i>URA3</i></b> encodes <i>Orotidine 5'-phosphate decarboxylase</i>—an enzyme that catalyzes one reaction in the synthesis of <i>pyrimidine ribonucleotides</i>. We obtained the <b><i>URA3</i></b> gene from the <p>PRS416</p> plasmid,which worked as a vector for our functional genes.</p> | ||
| + | |||
| + | |||
| + | <h4>Principle of operation</h4> | ||
| + | <h5>1) Pyrimidine biosynthetic pathway of<i> S. cerevisiae</i></h5> | ||
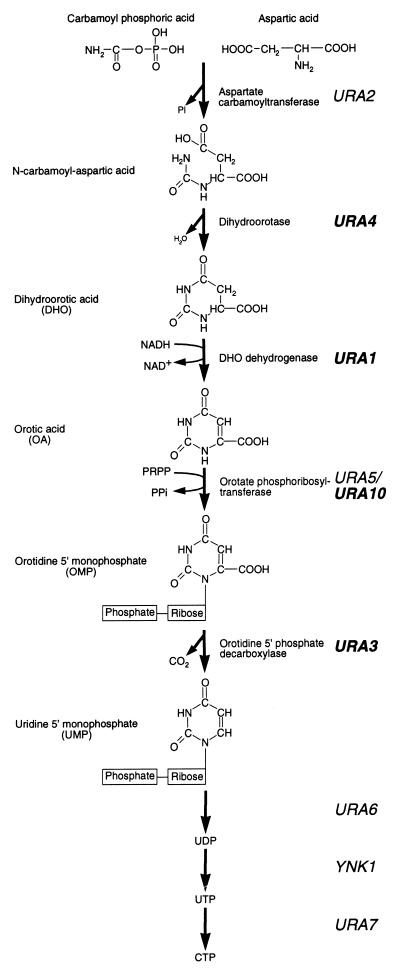
| + | <p>In Saccharomyces cerevisiae, the biosynthesis of pyrimidines involves the de novo synthesis of <i>UMP</i> from <i>glutamine</i>. <i>Carbamoyl phosphate</i>, derived from <i>glutamine</i>, undergoes a condensation reaction with <i>aspartic acid</i>, resulting in the formation of <i>N-carbamoyl aspartic acid</i>. Both the formation and subsequent condensation of <i>carbamoyl phosphate</i> are performed by <i>Ura2p</i>. The <i>pyrimidine</i> ring of <i>N-carbamoyl aspartic acid</i> is closed by the elimination of water to form <i>dihydroorotic acid (DHO)</i>, which is subsequently <i>oxidized</i> to form <i>orotic acid (OA)</i>, and a <i>ribose-phosphate</i> group is then added to form orotidine <i>5′-monophosphate (OMP)</i>. The formation of <i>OMP</i> is performed by two <i>isoenzymes</i>, <i>Ura5p</i> and <i>Ura10p</i>. <i>OMP</i> is then <i>decarboxylated</i> to yield <i>UMP</i>, which may subsequently be processed to form other <i>pyrimidines</i>. Regulation of this pathway occurs at several levels. First, <i>UTP</i> down-regulates the enzymatic activity of <i>Ura2p<i> and transcription of the <i>URA2 </i>gene. Second, under conditions of <i>pyrimidine</i> starvation, transcription of the <i>URA1, <b>URA3</b>, URA4</i>, and <i>URA10</i> genes (the <i>URA</i> genes) is increased some three- to eightfold. This increase in transcription is dependent on a transcriptional activator, <i>Ppr1p</i>.</p> | ||
| + | |||
| + | |||
| + | [[File:Tianjin-ura1.jpg|1400px|thumb|center|'''Fig 1'''.The <i>pyrimidine</i> metabolism pathway in yeasts. ]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <h5>2) Usage in yeast research</h5> | ||
| + | <p>The <b><i>URA3</i></b> gene in the yeasts used in lab has already been deleted. Hence the loss of <i>ODCase</i> activity leads to a lack of cell growth unless <i>uracil</i> or <i>uridine</i> is added to the media. The presence of the <b><i>URA3</i></b> gene in yeast restores <i>ODCase</i> activity, facilitating growth on media not supplemented with <i>uracil</i> or <i>uridine</i>, thereby allowing selection for yeast carrying the gene. In contrast, if <i>5-FOA (5-Fluoroorotic acid)</i> is added to the media, the active <i>ODCase</i> will convert <i>5-FOA</i> into the toxic compound (a suicide inhibitor) <i>5-fluorouracil</i> causing cell death, which allows for selection against yeast carrying the gene. </p> | ||
| + | <p>Since <b><i>URA3</i></b> allows for both positive and negative selection, it has been developed as a genetic marker for DNA transformations and other genetic techniques in bacteria and many fungal species. It is one of the most important genetic markers in yeast genetic modification. While <b><i>URA3</i></b> is a powerful selectable marker it has a high background. This background is because cells that pick up mutations in <b><i>URA3</i></b> may also grow on <i>5-FOA</i>. Colonies should be verified by a second assay such as PCR to confirm the desired strain has been created. </p> | ||
| + | |||
| + | |||
| + | <h4>Characterization</h4> | ||
| + | <h5>Positive selection</h5> | ||
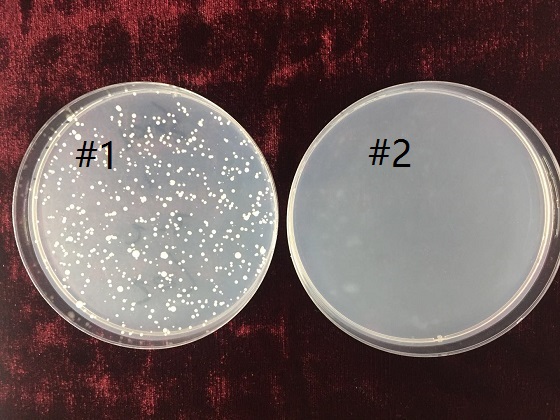
| + | <p>For characterizing the positive selecting function of <b><i>URA3</i></b> part, we designed the following experiment:</p> | ||
| + | <p>1. Incubate 2 test tubes of <i>yeasts-BY4741<i>, and numbered as #1, #2. (cultured in YPD, 30℃,6 months)</p> | ||
| + | <p>2. After incubation, the yeasts in #1 tube are transformed by <i>PRS416</i> plasmid, which contains URA3 gene as report gene. </p> | ||
| + | <p>3. Two groups of yeasts are spread on two <i>Sc-URA</i> plates, and hatch in the 30℃ incubator for 48 hr. </p> | ||
| + | <p>4. Examine the growth situation of yeasts on both plates. </p> | ||
| + | |||
| + | [[File:Tianjin-ura2.jpg|700px|thumb|center|'''Fig 2''' The plasmid profile for <i>PRS416</i> ]] | ||
| + | [[File:Tianjin-ura5.jpg|700px|thumb|center|'''Fig 3''' The result of our experiment to characterize the <b><i>URA3</i></b> gene for positive selection.]] | ||
| + | <p>Showed in the photograph above, yeasts in group 1#, with <i>PRS416</i> plasmid, has proliferated on the <i>Sc-URA</i> plate. Meanwhile, the other group without <b><i>URA3</i></b> gene are not able to grow on the <i>Sc-URA</i> plate. The result indicates the <b><i>URA3</i></b> gene is necessary for our yeasts (<i>BY4741</i>) to grow and proliferate on media not supplemented with uracil, thereby allowing selection for yeast carrying the gene. </p> | ||
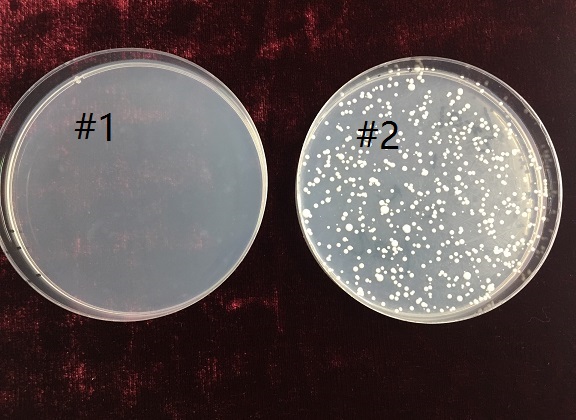
| + | <h5>negative selection</h5> | ||
| + | <p>In order to characterize the negative selecting function of <b><i>URA3</i></b> gene, after the transformation of #1 yeasts, we also spread the yeasts of group 1 on a <i>5-FOA</i> plate, and spread group 2 yeasts on another <i>5-FOA</i> plate in the meantime as control.</p> | ||
| + | [[File:Tianjin-ura6.jpg|700px|thumb|center|'''Fig 4''' The result of our experiment to characterize the <b><i>URA3</i></b> gene for negative selection.]] | ||
| + | <p>As presented above, yeasts #1(with <b><i>URA3</i></b> gene) haven’t amplified in the <i>5-FOA</i> plate, while yeasts #2 can proliferate on it, which testified the negative selecting function of <b><i>URA3</i></b> gene.</p> | ||
| + | <h4>Reference</h4> | ||
| + | <p>[1] Wikipedia-URA3 gene. https://en.wikipedia.org/wiki/URA3.</p> | ||
| + | <p>[2] Flynn, P. J.; Reece, R. J. (1999). "Activation of transcription by metabolic intermediates of the pyrimidine biosynthetic pathway". Molecular and Cellular Biology. 19.</p> | ||
| + | <hr> | ||
| + | <p>Actually, we built this new part based on Visit <partinfo>BBa_K319036</partinfo>. We supplemented more specific information concerning URA3 gene, and we accomplished two complete experiments to characterize its function as gene marker.</p> | ||
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | + | ||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
Latest revision as of 16:03, 27 October 2017
Ura3 gene
This is the part regulatory region from the URA3 gene coding for OMP decarboxylase, an essential protein in the uracil synthesis pathway in S. cerevisiae budding yeast. It is widely used as a nutrition tag in Saccharomyces cerevisiae.
Description
URA3, a gene on chromosome V in Saccharomvces cerevisiae, is widely used in researches concerning yeasts as a “marker gene” (systematic name YEL021W. URA3) and used as a label for chromosomes orplasmids. URA3 encodes Orotidine 5'-phosphate decarboxylase—an enzyme that catalyzes one reaction in the synthesis of pyrimidine ribonucleotides. We obtained the URA3 gene from the <p>PRS416
plasmid,which worked as a vector for our functional genes.</p>
Principle of operation
1) Pyrimidine biosynthetic pathway of S. cerevisiae
In Saccharomyces cerevisiae, the biosynthesis of pyrimidines involves the de novo synthesis of UMP from glutamine. Carbamoyl phosphate, derived from glutamine, undergoes a condensation reaction with aspartic acid, resulting in the formation of N-carbamoyl aspartic acid. Both the formation and subsequent condensation of carbamoyl phosphate are performed by Ura2p. The pyrimidine ring of N-carbamoyl aspartic acid is closed by the elimination of water to form dihydroorotic acid (DHO), which is subsequently oxidized to form orotic acid (OA), and a ribose-phosphate group is then added to form orotidine 5′-monophosphate (OMP). The formation of OMP is performed by two isoenzymes, Ura5p and Ura10p. OMP is then decarboxylated to yield UMP, which may subsequently be processed to form other pyrimidines. Regulation of this pathway occurs at several levels. First, UTP down-regulates the enzymatic activity of Ura2p<i> and transcription of the <i>URA2 gene. Second, under conditions of pyrimidine starvation, transcription of the URA1, URA3, URA4, and URA10 genes (the URA genes) is increased some three- to eightfold. This increase in transcription is dependent on a transcriptional activator, Ppr1p.
2) Usage in yeast research
The URA3 gene in the yeasts used in lab has already been deleted. Hence the loss of ODCase activity leads to a lack of cell growth unless uracil or uridine is added to the media. The presence of the URA3 gene in yeast restores ODCase activity, facilitating growth on media not supplemented with uracil or uridine, thereby allowing selection for yeast carrying the gene. In contrast, if 5-FOA (5-Fluoroorotic acid) is added to the media, the active ODCase will convert 5-FOA into the toxic compound (a suicide inhibitor) 5-fluorouracil causing cell death, which allows for selection against yeast carrying the gene.
Since URA3 allows for both positive and negative selection, it has been developed as a genetic marker for DNA transformations and other genetic techniques in bacteria and many fungal species. It is one of the most important genetic markers in yeast genetic modification. While URA3 is a powerful selectable marker it has a high background. This background is because cells that pick up mutations in URA3 may also grow on 5-FOA. Colonies should be verified by a second assay such as PCR to confirm the desired strain has been created.
Characterization
Positive selection
For characterizing the positive selecting function of URA3 part, we designed the following experiment:
1. Incubate 2 test tubes of yeasts-BY4741<i>, and numbered as #1, #2. (cultured in YPD, 30℃,6 months)</p> <p>2. After incubation, the yeasts in #1 tube are transformed by <i>PRS416 plasmid, which contains URA3 gene as report gene.
3. Two groups of yeasts are spread on two Sc-URA plates, and hatch in the 30℃ incubator for 48 hr.
4. Examine the growth situation of yeasts on both plates.
Showed in the photograph above, yeasts in group 1#, with PRS416 plasmid, has proliferated on the Sc-URA plate. Meanwhile, the other group without URA3 gene are not able to grow on the Sc-URA plate. The result indicates the URA3 gene is necessary for our yeasts (BY4741) to grow and proliferate on media not supplemented with uracil, thereby allowing selection for yeast carrying the gene.
negative selection
In order to characterize the negative selecting function of URA3 gene, after the transformation of #1 yeasts, we also spread the yeasts of group 1 on a 5-FOA plate, and spread group 2 yeasts on another 5-FOA plate in the meantime as control.
As presented above, yeasts #1(with URA3 gene) haven’t amplified in the 5-FOA plate, while yeasts #2 can proliferate on it, which testified the negative selecting function of URA3 gene.
Reference
[1] Wikipedia-URA3 gene. https://en.wikipedia.org/wiki/URA3.
[2] Flynn, P. J.; Reece, R. J. (1999). "Activation of transcription by metabolic intermediates of the pyrimidine biosynthetic pathway". Molecular and Cellular Biology. 19.
Actually, we built this new part based on Visit BBa_K319036. We supplemented more specific information concerning URA3 gene, and we accomplished two complete experiments to characterize its function as gene marker.
Usage and Biology
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 669
Illegal SapI.rc site found at 516