Difference between revisions of "Part:BBa K1936001"
Mdickmanns (Talk | contribs) (→Usage & Biology) |
Mdickmanns (Talk | contribs) (→Characterization) |
||
| (11 intermediate revisions by 2 users not shown) | |||
| Line 20: | Line 20: | ||
We chose Tom5 from <html><i>Saccharomyces cerevisiae</i></html> as an OMM anchor domain for our construct. It shows high structural similarity to its mammalian homologues. [3] Due to its relatively short cDNA sequence of 153bp we have favored it over other species orthologs, since our need for minimal size constructs is of major importance for the delivery of DNA via viral vectors. In the process of regular cloning as a basic experimental approach in cell culture small plasmid-size also increases the efficiency of transfection. | We chose Tom5 from <html><i>Saccharomyces cerevisiae</i></html> as an OMM anchor domain for our construct. It shows high structural similarity to its mammalian homologues. [3] Due to its relatively short cDNA sequence of 153bp we have favored it over other species orthologs, since our need for minimal size constructs is of major importance for the delivery of DNA via viral vectors. In the process of regular cloning as a basic experimental approach in cell culture small plasmid-size also increases the efficiency of transfection. | ||
| − | The LOV domain originated from <html><i>Avena sativa</i></html>. The T406A,T407A double mutation has been shown to stabilize the N-terminal A′α helix of AsLOV2 which reduces the dark activity of J<html>α</html> helix binding while also increasing its docking affinity to ePDZ. [2] | + | The LOV domain originated from <html><i>Avena sativa</i></html>. The T406A,T407A double mutation we used has been shown to stabilize the N-terminal A′α helix of AsLOV2 which reduces the dark activity of J<html>α</html> helix binding while also increasing its docking affinity to ePDZ. [2] |
Due to the C-terminal position of the J<html>α</html> helix Tom5 and eGFP were fused to the N-terminal side of LOV2 peptide. | Due to the C-terminal position of the J<html>α</html> helix Tom5 and eGFP were fused to the N-terminal side of LOV2 peptide. | ||
| Line 87: | Line 87: | ||
</html> | </html> | ||
| − | < | + | ===Characterization=== |
| + | <html><h2>Flourescence imaging of constitutive expression of K1936001 and K1936002</h2><html> | ||
| + | <html><img src="https://static.igem.org/mediawiki/2016/2/2d/T--duesseldorf--fig3.png" style="max-width:100%;"><br /><br></html> | ||
| + | <html><h2>STED microscopy comparison of expression levels depending on illumination</h2></html> | ||
| + | <html><img src="https://static.igem.org/mediawiki/2016/2/2e/T--duesseldorf--mikroskopimg.png" style="max-width:100%;"><br /><br> </html> | ||
| + | <html><h2>relative mCherry production depending on illumination</h2></html> | ||
| + | <html><img src="https://static.igem.org/mediawiki/2016/0/04/T--duesseldorf--fig.png" style="max-width:100%;"><br /><br></html> | ||
| + | |||
| + | <html><b>Plasmids:</b><br></html> | ||
| + | 15 - tetO + <html><a href="https://parts.igem.org/Part:BBa_K1936000">K1936000</a><br></html> | ||
| + | 022 - tetR+PIF6<html><br></html> | ||
| + | LX - K1936001 with constitutive expression<html><br></html> | ||
| + | PX - <html><a href="https://parts.igem.org/Part:BBa_K1936000">K1936000</a></html> with constitutive expression<html><br><br></html> | ||
| + | Check our <html><a href="http://2016.igem.org/Team:Duesseldorf/Description">Wiki</a></html> for further information on those constructs. | ||
===References=== | ===References=== | ||
Latest revision as of 21:53, 29 October 2016
Tom5-eGFP-LOV2
Usage & Biology
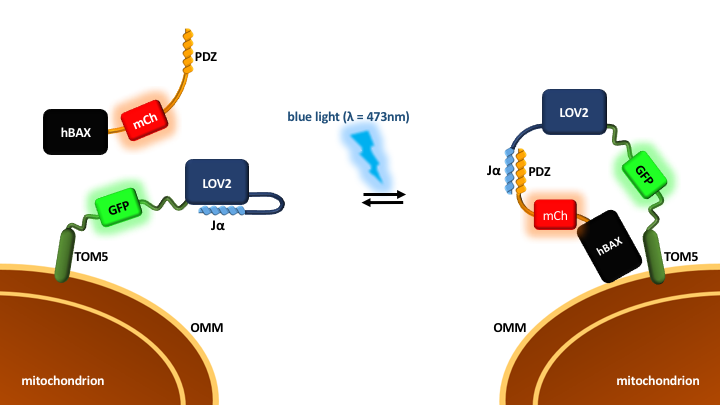
While LOV2 domain was previously described for optogenetical control of gene expression (BBa_K1742000) and as reporter gene (BBa_K660003), we took a new approach. The described fusion protein Tom5-eGFP-LOV2 is designed to recruit proteins of choice to the outer mitochondrial membrane (OMM) in engineered eukaryotic cells.
Tom5 is a part of the eukaryotic translocase of outer membrane protein complex and has a single membrane anchor unit that integrates into OMM. [1] We fused it to the N-terminal end of LOV2 domain from Avena sativa. Upon exposure to blue light (λ=473nm) the LOV2 peptide undergoes a conformational change and exposes its C-terminal Jα helix which is then able to bind a specifically engineered PDZ domain (BBa_K1470005). [2] The fused enhanced green fluorenscence protein (eGFP) allows the tracking of cellular localization by microscopy.
Our blue light dependent system can be used to recruit any protein of interest to the outer mitochondrial membrane by simply fusing it to an ePDZ domain.
For our project we utilized the light dependent OMM recruitment as a potential method for cancer treatment. As shown in Fig.1 it recruits BAX protein which is fused to the ePDZ domain (see BBa_K1936000). Once BAX is localized at the mitochondrial membrane it leads to formation of apoptotic channels. Through those channels cytochrome c can be released from the mitochondrial intermembrane space to the cytosol which triggers apoptosome formation. This allows us to kill abnormal cells with attainment of a very high level of spatiotemporal specificity compared to traditional cancer therapies (http://2016.igem.org/Team:Duesseldorf).
Design
We chose Tom5 from Saccharomyces cerevisiae as an OMM anchor domain for our construct. It shows high structural similarity to its mammalian homologues. [3] Due to its relatively short cDNA sequence of 153bp we have favored it over other species orthologs, since our need for minimal size constructs is of major importance for the delivery of DNA via viral vectors. In the process of regular cloning as a basic experimental approach in cell culture small plasmid-size also increases the efficiency of transfection.
The LOV domain originated from Avena sativa. The T406A,T407A double mutation we used has been shown to stabilize the N-terminal A′α helix of AsLOV2 which reduces the dark activity of Jα helix binding while also increasing its docking affinity to ePDZ. [2] Due to the C-terminal position of the Jα helix Tom5 and eGFP were fused to the N-terminal side of LOV2 peptide.
Both linkers were designed to be flexibel by using short sequences of amino acids G, S & A. The flexibility is especially important for bringing the ePDZ-bound protein close to the membrane. [4] The exact amino acid sequences are shown in Tabel 1.
| Linker | Sequence |
| L1 | GGGS-GGGGS-GGGS |
| L2 | GSGAAGSGAG |
The full sequence except for the two linkers has been codon optimized for expression in H.sapiens by using the IDT optimization tool and where then synthesized as gBlock.
| Name | Start | Stop | Length [bp] |
| Tom5 | 1 | 153 | 153 |
| Linker1 () | 154 | 192 | 39 |
| eGFP | 193 | 906 | 714 |
| Linker2 | 907 | 936 | 30 |
| LOV2 | 937 | 1368 | 432 |
Characterization
Flourescence imaging of constitutive expression of K1936001 and K1936002

STED microscopy comparison of expression levels depending on illumination

relative mCherry production depending on illumination

Plasmids:
15 - tetO + K1936000
022 - tetR+PIF6
LX - K1936001 with constitutive expression
PX - K1936000 with constitutive expression
Check our Wiki for further information on those constructs.
References
[1] Dietmeier K, Hönlinger A, Bömer U, Dekker PJ, Eckerskorn C, Lottspeich F, Kübrich M, Pfanner N (1997) Tom5 functionally links mitochondrial preprotein receptors to the general import pore. Nature. 1997 Jul 10;388(6638):195-200.
[2] Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M (2012) TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012 Mar 4;9(4):379-84. doi: 10.1038/nmeth.1904.
[3] Kato H, Mihara K (2008) Identification of Tom5 and Tom6 in the preprotein translocase complex of human mitochondrial outer membrane. Biochem Biophys Res Commun. 2008 May 9;369(3):958-63. doi: 10.1016/j.bbrc.2008.02.150. Epub 2008 Mar 10.
[4] Chen X1, Zaro JL, Shen WC (2013) Fusion protein linkers: property, design and functionality. Adv Drug Deliv Rev. 2013 Oct;65(10):1357-69. doi: 10.1016/j.addr.2012.09.039. Epub 2012 Sep 29.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 33
Illegal BamHI site found at 712
Illegal XhoI site found at 515 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1226