Difference between revisions of "Part:BBa K1893007"
| (One intermediate revision by the same user not shown) | |||
| Line 8: | Line 8: | ||
Quorum sensing is a naturally occurring mechanism that certain strains of bacteria use to regulate gene expression in response to their population density. These bacteria secrete autoinducer signalling molecules, such as N-acyl homoserine lactones (AHLs), that bind to transcription factors to alter gene expression. | Quorum sensing is a naturally occurring mechanism that certain strains of bacteria use to regulate gene expression in response to their population density. These bacteria secrete autoinducer signalling molecules, such as N-acyl homoserine lactones (AHLs), that bind to transcription factors to alter gene expression. | ||
| − | In this case, the constitutively expressed CinR transcriptional regulator (K1893028) is activated by the binding of 3O-C14 AHL, an AHL quorum signal. The activated CinR regulator binds to a CinR-inducible promoter, pCin, upstream of a GFP reporter. As a result, GFP is expressed when the receiver device is induced with 3O-C14 AHL. We designed this part to characterize the activation range of the Cin receiver device. We also characterised the cross-talk of the Cin receiver device with other quorum-sensing systems, in order to determine its orthogonality. | + | In this case, the constitutively expressed CinR transcriptional regulator [https://parts.igem.org/Part:BBa_K1893028 (K1893028)] is activated by the binding of 3O-C14 AHL, an AHL quorum signal. The activated CinR regulator binds to a CinR-inducible promoter, pCin, upstream of a GFP reporter. As a result, GFP is expressed when the receiver device is induced with 3O-C14 AHL. We designed this part to characterize the activation range of the Cin receiver device. We also characterised the cross-talk of the Cin receiver device with other quorum-sensing systems, in order to determine its orthogonality. |
===Characterisation data=== | ===Characterisation data=== | ||
| Line 16: | Line 16: | ||
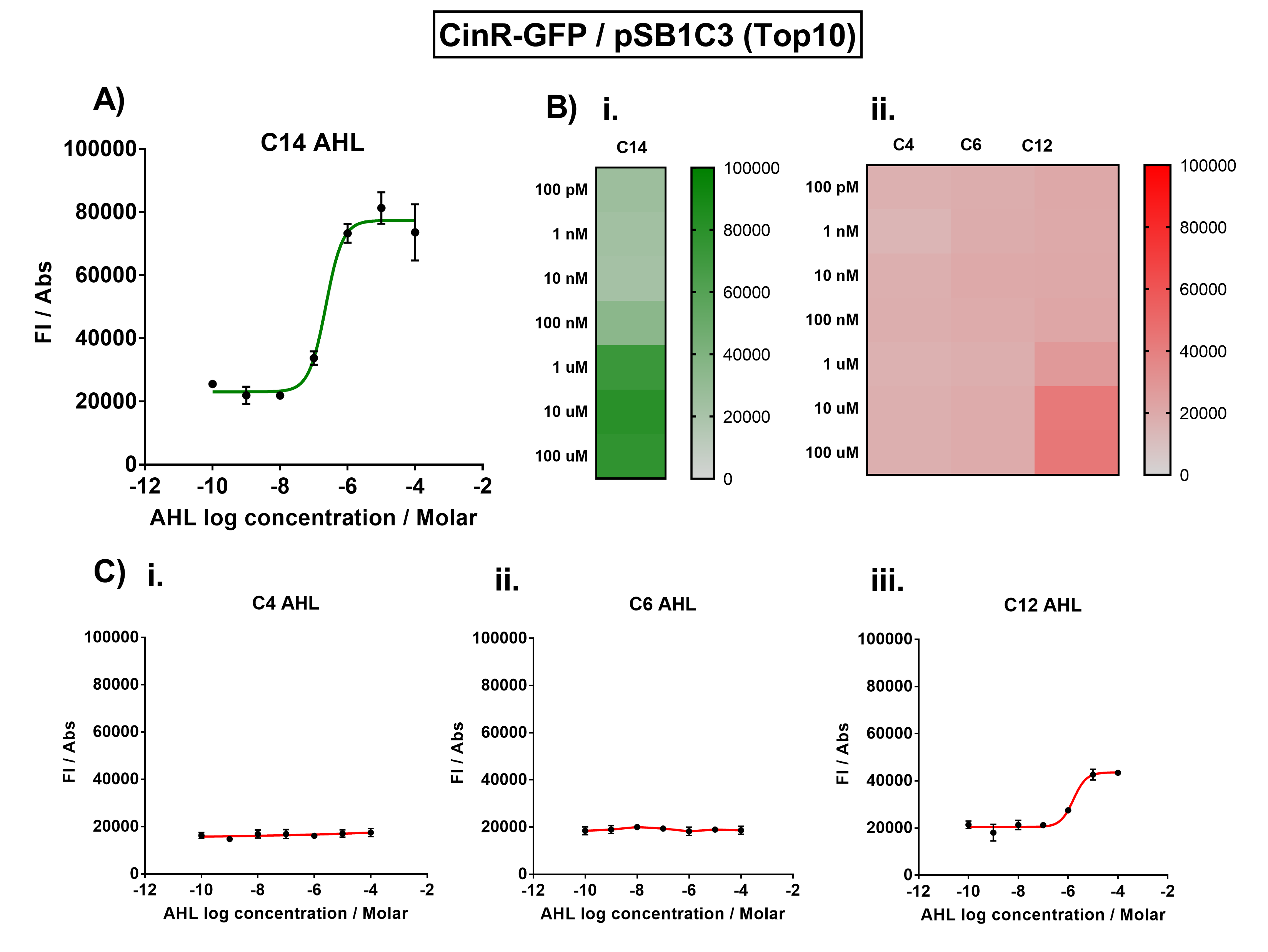
[[File:IC16_CinGraph.png|700px|center|]] | [[File:IC16_CinGraph.png|700px|center|]] | ||
| − | Figure 1. Characterisation of the Cin response device (BBa_K1893007). (A) Transfer function curve of normalised fluorescence against cognate inducer C14-AHL concentrations. (B) Heat map of normalised fluorescence of CinR-GFP system over a range of AHL concentrations: (i) Binding of CinR-GFP to its cognate AHL (3O-C14 AHL). (ii) Binding of RhlR-GFP to 3 non-cognate AHLs (C4 AHL, 3O-C6 AHL, 3O-C12 AHL). (C) Transfer function curves of normalised fluorescence against non-cognate inducer AHL (3O-C14 AHL) concentrations to investigate inducer AHL crosstalk: (i) C4-AHL (C4 AHL) of the Rhl system (ii) C6-AHL (3O-C6 AHL) of the Lux system (iii) C12-AHL (3O-C12 AHL) of the Las system. Experiments were performed in E. coli Top10 cell strain cultured at 37°C. Normalised fluorescence was calculated by dividing fluorescent signal by cell density (OD600). Fluorescence measurements were recorded at 180 minutes. Reported values represent the mean normalised fluorescence value from 3 technical repeats and error bars represent standard deviation of these. | + | Figure 1. Characterisation of the Cin response device [https://parts.igem.org/Part:BBa_K1893007 (BBa_K1893007)]. (A) Transfer function curve of normalised fluorescence against cognate inducer C14-AHL concentrations. (B) Heat map of normalised fluorescence of CinR-GFP system over a range of AHL concentrations: (i) Binding of CinR-GFP to its cognate AHL (3O-C14 AHL). (ii) Binding of RhlR-GFP to 3 non-cognate AHLs (C4 AHL, 3O-C6 AHL, 3O-C12 AHL). (C) Transfer function curves of normalised fluorescence against non-cognate inducer AHL (3O-C14 AHL) concentrations to investigate inducer AHL crosstalk: (i) C4-AHL (C4 AHL) of the Rhl system (ii) C6-AHL (3O-C6 AHL) of the Lux system (iii) C12-AHL (3O-C12 AHL) of the Las system. Experiments were performed in E. coli Top10 cell strain cultured at 37°C. Normalised fluorescence was calculated by dividing fluorescent signal by cell density (OD600). Fluorescence measurements were recorded at 180 minutes. Reported values represent the mean normalised fluorescence value from 3 technical repeats and error bars represent standard deviation of these. |
===Analysis=== | ===Analysis=== | ||
Latest revision as of 20:45, 28 October 2016
Cin receiver with GFP (CinR+pCin+GFP)
This composite part is based on the Cin quorum-sensing system from Rhizobium leguminosarum. It consists of a Cin receiver device and a GFP reporter that is activated in the presence of a 3O-C14 AHL signal.
Usage and Biology
Quorum sensing is a naturally occurring mechanism that certain strains of bacteria use to regulate gene expression in response to their population density. These bacteria secrete autoinducer signalling molecules, such as N-acyl homoserine lactones (AHLs), that bind to transcription factors to alter gene expression.
In this case, the constitutively expressed CinR transcriptional regulator (K1893028) is activated by the binding of 3O-C14 AHL, an AHL quorum signal. The activated CinR regulator binds to a CinR-inducible promoter, pCin, upstream of a GFP reporter. As a result, GFP is expressed when the receiver device is induced with 3O-C14 AHL. We designed this part to characterize the activation range of the Cin receiver device. We also characterised the cross-talk of the Cin receiver device with other quorum-sensing systems, in order to determine its orthogonality.
Characterisation data
The activation range of CinR by its cognate inducer (3O-C14 AHL) was characterised in TOP10 E.Coli cells. Cells transformed with the Cin response device were cultured to the exponential phase and treated with appropriate concentrations of 3O-C14 AHL in 96-well microplates. These induced cells were grown in the microplates and their fluorescence and absorbance values (OD 600) were monitored over time using a microplate reader. The reported values for the normalised fluorescence represents the values recorded 180 minutes after AHL induction. The normalised fluorescence was calculated by dividing fluorescence values by absorbance values and correcting for LB autofluorescence.
In order to characterise the orthogonality of the Cin system, we measured absorbance and fluorescence of the cells in the plate reader after treating them with varying concentrations of 3 different AHL signals (C4 AHL of the Rhl system, 3O-C6 AHL of the Lux system, and 3O-C12 AHL of the Las system). This allowed us to determine whether these non-cognate AHLs were capable of activating the CinR response protein, and therefore the level of crosstalk between the quorum sensing systems.
Figure 1. Characterisation of the Cin response device (BBa_K1893007). (A) Transfer function curve of normalised fluorescence against cognate inducer C14-AHL concentrations. (B) Heat map of normalised fluorescence of CinR-GFP system over a range of AHL concentrations: (i) Binding of CinR-GFP to its cognate AHL (3O-C14 AHL). (ii) Binding of RhlR-GFP to 3 non-cognate AHLs (C4 AHL, 3O-C6 AHL, 3O-C12 AHL). (C) Transfer function curves of normalised fluorescence against non-cognate inducer AHL (3O-C14 AHL) concentrations to investigate inducer AHL crosstalk: (i) C4-AHL (C4 AHL) of the Rhl system (ii) C6-AHL (3O-C6 AHL) of the Lux system (iii) C12-AHL (3O-C12 AHL) of the Las system. Experiments were performed in E. coli Top10 cell strain cultured at 37°C. Normalised fluorescence was calculated by dividing fluorescent signal by cell density (OD600). Fluorescence measurements were recorded at 180 minutes. Reported values represent the mean normalised fluorescence value from 3 technical repeats and error bars represent standard deviation of these.
Analysis
The results from Figure 1A show that the concentration range of AHL (3O-C14 AHL) required for the activation of the Cin response device was 10nM-100uM. Furthermore, it can be seen from Figure 1C that CinR does not appear to be activated by C4 AHL or 3O-C6 AHL, suggesting that the Cin quorum system is orthogonal with Rhl and Lux. However, it is seen that CinR is activated by 3O-C12 AHL, suggesting that the Cin and Las quorum systems lack orthogonality.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 611
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1698