Difference between revisions of "Part:BBa K2127001"
| Line 22: | Line 22: | ||
[[File:Team_Ingenuity_Lab_Psbb_Gel.png]] | [[File:Team_Ingenuity_Lab_Psbb_Gel.png]] | ||
| − | + | ==Funtionality== | |
| + | |||
| + | The composit part BBa_K2127001 was also tested for functionality. The lac promotor allows for inducement using IPTG and the His tag allows for purification of the protein. The figure shows that expected band of our CP47His protein at 48kDa. Since lac is a leaky promoter the darkening of the band in the cell lysate is expected. The flow through and the washes also contain our band wih increasing clarity. The inability for the his-tag to bind properly to the column was due to the use of a protein inhibitor cocktail that contained EDTA. The EDTA caused chelation with the nickel ligands and resulted in our protein washing out with subsequent washes. The gel does confirm the the BBa_K2127001 is being successfuly expressed according to our design. Further work is needs to confirm the difference in expression levels between BBa_K2127001 and BBa_K2127002. | ||
| + | |||
| + | |||
| + | [[File:Team_Ingenuity_Lab_TFBSexpression.png|thumb|500px|center| <strong>Figure 2</strong> DH5aF'Iq cells transformed with the shipping vector plus our BBa_K2127001 composite part plasmid were induced using 400mM IPTG for 1.5 hours. Expression was carried out at 37 degrees Celsius for 1.5 hours. Cells were chemically lysed under denaturing conditions. Lysate was then being incubated for 1 hour with Ni-NTA beads. The slurry was then packed into a gravity column and beads were washed with 20mM imidiazole wash buffer and eluted in 250mM imidizaole elution buffer. Samples were then collected and run on 12% SDS-PAGE at 120 V for 45 min]] | ||
Revision as of 22:47, 27 October 2016
Inducible High Expression His-Tagged Photosystem II CP47 subunit
Cyanobacteria is an not optimal system to manipulate the protein expression mainly due of the lack of strong promoters. Pcpc560 is a Cyanobacterial super promoter discovered by Dr. Ma’s team that contains two predicted promoter sites and a transcription binding site containing 14 homologous bacterial regions upstream of the regulated cpcB gene. Using Pcpc560, Dr. Ma’s team demonstrated that functional protein was produced at 15% of the total cell protein volume. Our team decided to test the effect of the transcription factor binding site on protein production in E.Coli cells DH5α. We believe that the predicted 14 homologous binding sites from Pcpc560 will allow us to increase the protein production greatly using the E.Coli DH5α. We constructed a biobrick containing the 14 transcription binding sites followed by a strong RBS (BBa_B0030) and a mutant HIS-Tag psbB gene. The super promoter can be an valuable asset to other iGEM teams for protein production and therefore we have decided to submit the Construct along with the Transcription Binding Site from Pcpc560.
We envision that this construct will be helpful in allowing teams to directly begin working with Photosystem components. Being sent as an expressible construct, teams will have the option of either using the construct directly for protein work, or isolating the genetic sequence and manipulating it to their needs.
Usage and Biology
The his-tagged CP47 subunit is approximately 48kDa and is a transmembrane protein. As part of the Photosystem II complex, the subunit binds chlorophyll and acts as part of the light capturing array. The subunit ligand sites allow for electron movement across the peptide to facilitate movement within the electron transfer chain from both the light capturing array and from the photolysis of water. The subunit also composes part of the manganese calcium complex for oxygen evolution. It is extremely integral to the function of the PSII complex. Also in conjunction to its functions, CP47 also forms part of the quinone channel which allows direct transfer of electrons from the D1 monomer to quinones of the electron transport chain
Verification
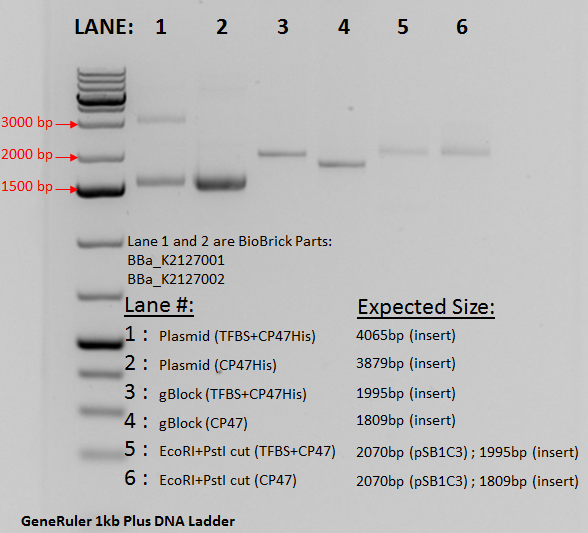
A plasmid containing the composite part was successfully made and transformed into E. coli DH5aF'Iq. The plasmid was extracted using miniprep and double digested using EcoRI and Psti. The gel below confirms the transformation. Lane 1 contains the undigested plasmid running at approximately 1500 bp due to a supercoiled nature. An unexpected band at 3500 bp is seen and could be due to dimerization of the pSB1C3 vector. Support of this is due to the disappearance of the band once the sample is digested as seen in Lane 5. Lane 3 contains the gBlock from IDT and acts as a size control. The band is seen aa expected at 1995 bp. In lane 5, the digested plasmid is seen. Due to both the pSB1C3 vector and the BBa_K2127001 insert being very close to each other at 1995bp and 2070 bp, the resolution of the gel did not allow for a clear distinction of the bands. Compared to Lane 1 it can been inferred that digestion did occur since the run time changes significantly between the linearized digest and the undigested supercoiled plasmid.
Funtionality
The composit part BBa_K2127001 was also tested for functionality. The lac promotor allows for inducement using IPTG and the His tag allows for purification of the protein. The figure shows that expected band of our CP47His protein at 48kDa. Since lac is a leaky promoter the darkening of the band in the cell lysate is expected. The flow through and the washes also contain our band wih increasing clarity. The inability for the his-tag to bind properly to the column was due to the use of a protein inhibitor cocktail that contained EDTA. The EDTA caused chelation with the nickel ligands and resulted in our protein washing out with subsequent washes. The gel does confirm the the BBa_K2127001 is being successfuly expressed according to our design. Further work is needs to confirm the difference in expression levels between BBa_K2127001 and BBa_K2127002.

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 658
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 782
Illegal AgeI site found at 1379
Illegal AgeI site found at 1583 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1103
Illegal SapI site found at 1780