Difference between revisions of "Part:BBa K1980009"
| (12 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K1980009 short</partinfo> | <partinfo>BBa_K1980009 short</partinfo> | ||
| − | + | ==Description== | |
| − | + | <p> This synthetic promoter system includes the response regulator CusR (part of the CusR/S copper sensitive two-component system found in <i>E. coli</i>) and the an RFP variant (mKate2) expressed downstream of a copper sensitive promoter (pCusC). As CusR is found downstream of the promoter it activates means that a positive feedback loop is created when sufficient copper is sensed in the environment.</p> | |
| − | + | <p>However we were unable to obtain of copy of this system without a point mutation which appears to have broken the feedback loop.</p> | |
| − | <p> This synthetic promoter system includes the response regulator CusR (part of the CusR/S copper sensitive two-component system found in <i>E. coli</i>) and the an RFP variant (mKate2) expressed downstream of a copper sensitive promoter (pCusC). | + | |
| − | + | ||
| − | + | ||
<!-- --> | <!-- --> | ||
| Line 15: | Line 12: | ||
==Usage and Biology== | ==Usage and Biology== | ||
| + | Our project was to investigate a probiotic treatment for the copper-accumulation disorder: Wilson's disease. This required a system able to detect dietary copper, ideally in the range over which copper concentration changes after a meal (around 5-10μM). | ||
| + | |||
<p> <i> E. coli </i> responds to copper is the CusS/CusR two-component system. This consists of the transmembrane histidine kinase enzyme: CusS in the bacterial cytoplasmic membrane and a cytoplasmic response regulator: CusR.</p> | <p> <i> E. coli </i> responds to copper is the CusS/CusR two-component system. This consists of the transmembrane histidine kinase enzyme: CusS in the bacterial cytoplasmic membrane and a cytoplasmic response regulator: CusR.</p> | ||
| Line 20: | Line 19: | ||
<p>In <i>E. coli</i> this box is present between the promoters for CusCFBA operon which encodes a multi-protein pump that exports cytoplasmic and periplasmic copper from the cell and the CusRS operon which encodes the two component system (therefore acting as a positive feedback loop in vivo).<sup>(1)</sup></p> | <p>In <i>E. coli</i> this box is present between the promoters for CusCFBA operon which encodes a multi-protein pump that exports cytoplasmic and periplasmic copper from the cell and the CusRS operon which encodes the two component system (therefore acting as a positive feedback loop in vivo).<sup>(1)</sup></p> | ||
| + | |||
| + | A similar feedback loop with CusR was shown to be more sensitive to copper by Ravikumar et al.<sup>(2)</sup> | ||
| + | |||
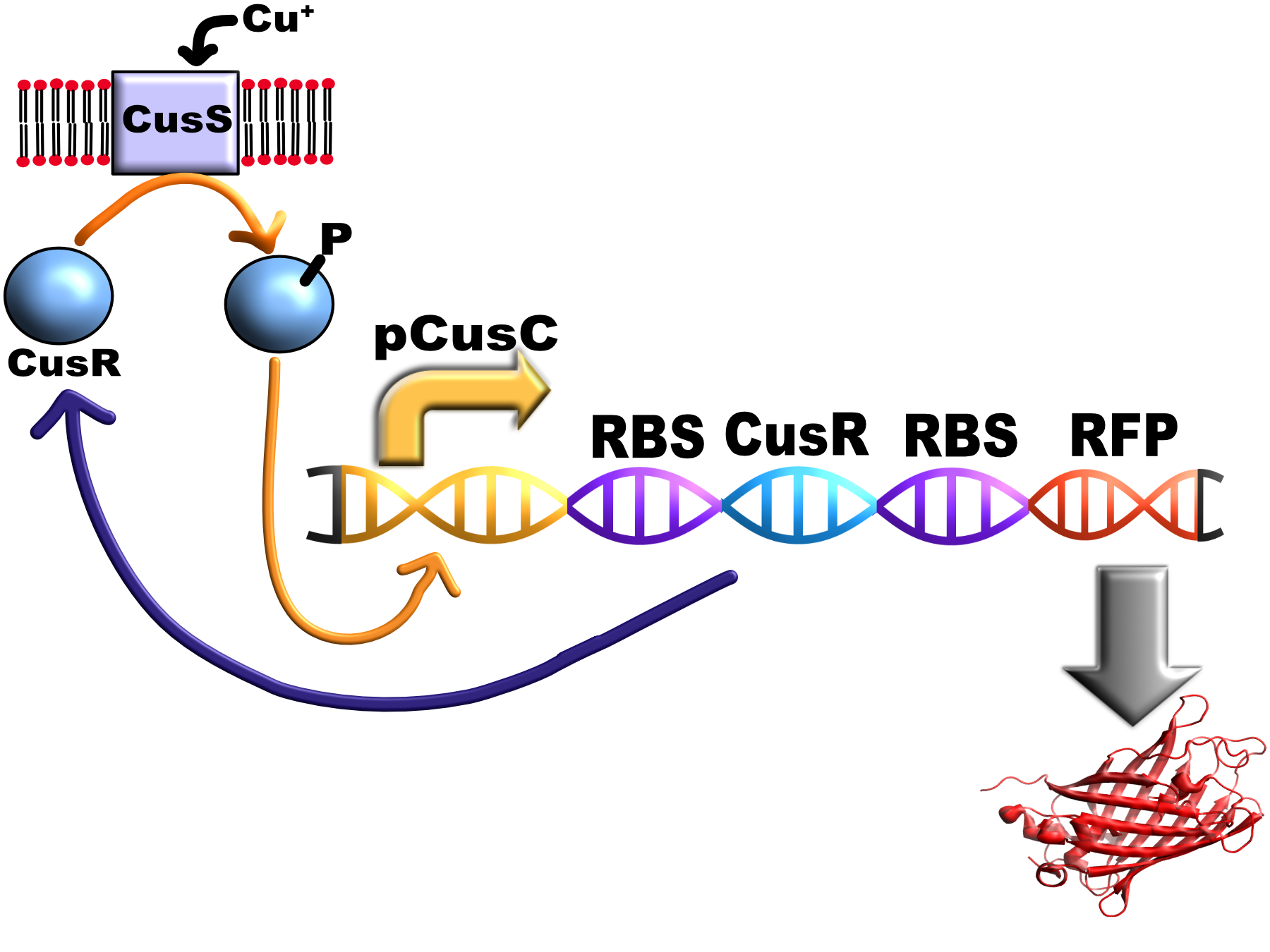
| + | <p> The following picture is a representation of how the construct works, theoretically <i>in vivo</i>:</p> | ||
| + | [[Image:T--Oxford--FCK_diagram_v2.jpg|400px|thumb|center|Schematic of the system]] | ||
==Experience== | ==Experience== | ||
| − | <p>We cloned pCusC CusR RFP from a gBlock into the shipping plasmid pSB1C3. <i>E. coli</i> strain MG1655 was transformed using the specific recombinant plasmid and a 5ml culture of a transformed colony was grown overnight. | + | <p>We cloned pCusC CusR RFP from a gBlock into the shipping plasmid pSB1C3. <i>E. coli</i> strain MG1655 was transformed using the specific recombinant plasmid and a 5ml culture of a transformed colony was grown overnight. Flow cytometry and plate reader experiments were performed to test sensitivity of the construct to various copper concentration</p> |
| − | + | ||
| − | |||
| − | [[Image:T--Oxford--FCKDATA.jpeg|400px|thumb| | + | ===Plate Reader=== |
| + | [[Image:T--Oxford--FCKDATA.jpeg|400px|thumb|right|The results of a plate reader experiment with different copper concentrations after four hours. Error bars show standard deviation of four repeats]] | ||
| + | <p> | ||
| + | We tested the pCusC promoter with the fluorescent protein mKate2 (a form of RFP excitation/emission maxima at 588 and 633 nm). | ||
| + | </p> | ||
| + | <p> | ||
| + | To account for the number of cells present at different copper concentrations and different times we measured the optical density (OD) as a proportional measure of the number of cells present. For our parts using RFP we measured the optical density at 700nm for these parts as measuring OD600 would get interference from the fluorescent protein. | ||
| + | </p> | ||
| + | <p> | ||
| + | A range of copper concentrations were prepared from stock solutions. A large volume plate was then prepared with 10μl of copper solution, 10μl of overnight culture and 980μl of broth with antibiotic. This resulted in a 1 in 100 dilution of the copper solutions prepared. | ||
| + | </p> | ||
| + | <p> | ||
| + | This large-volume plate was then centrifuged to mix the solutions and then 200μl transferred to a small-volume plate with a clear lid and then placed in the plate reader. </p> | ||
| + | <p> | ||
| + | Four biological repeats of the part to be tested in MG1655 at ten different copper concentrations were measured as well as four repeats of a MG1655 negative control. | ||
| + | </p> | ||
| + | <p> | ||
| + | The plate reader measured the fluorescence and OD every ten minutes for at least 12 hours, shaking between measurements. | ||
| + | </p> | ||
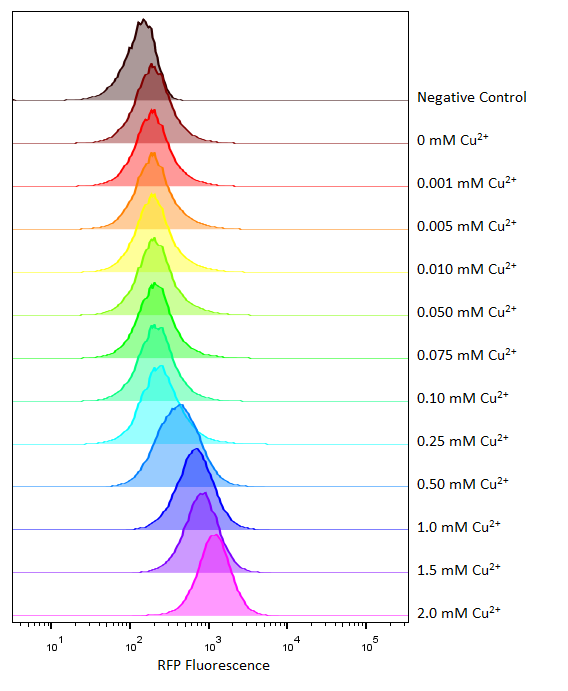
| − | [[Image:T--Oxford--FCKDATAcytometry.jpeg|400px|thumb| | + | ===Flow cytometry=== |
| + | [[Image:T--Oxford--FCKDATAcytometry.jpeg|400px|thumb|right|The population distribution of this part at different copper concentrations as measured by flow cytometry]] | ||
| + | <p> | ||
| + | Flow cytometry is a technique whereby cells are passed individually into the path of a beam of light. The frequency range of incident light can be adjusted to allow excitation of specific fluorophores in the sample of cells. Downstream detectors can measure fluorescent emission, and this data can be used to quantify the amount of fluorophore in each cell.</p> | ||
| + | <p>To ensure comparability of experiments, all cells were grown for 3-4hrs (until entrance into the exponential growth phase) at 37°C and 225rpm shaking in the presence of the inducer before measurement. This allowed adequate time for activation of expression by the promoter systems. The negative control used in all cases were MG1655 bacteria containing an empty shipping plasmid. As these did not contain any fluorescent molecules, this population could be used to set the negative “gate” (i.e. the background fluorescence of the bacterial cells). Although the experiments were tedious as every sample had to be measured manually, the results were of remarkably high quality, were clearly interpretable, and fit very well with the other experimental data. | ||
| + | </p> | ||
| + | |||
| + | ===Conclusions=== | ||
| + | <p>This construct was an attempt to make a positive feedback system based upon pCusC. However, after many attempts we only managed to obtain a version with a point mutation (Val to Ala) in the CusR DNA-binding domain. The tests indicate that the mutation was tolerable but found no evidence of a copper sensitive system.</p> | ||
| Line 36: | Line 64: | ||
<p> (2) Sambandam Ravikumar, Van Dung Pham, Seung Hwan Lee, Ik-keun Yoo, Soon Ho Hong (2012) “Modification of CusSR bacterial two-component systems by the introduction of an inducible positive feedback loop” Journal of Industrial Microbiology & Biotechnology June 2012, Volume 39, Issue 6, pp 861–868</p> | <p> (2) Sambandam Ravikumar, Van Dung Pham, Seung Hwan Lee, Ik-keun Yoo, Soon Ho Hong (2012) “Modification of CusSR bacterial two-component systems by the introduction of an inducible positive feedback loop” Journal of Industrial Microbiology & Biotechnology June 2012, Volume 39, Issue 6, pp 861–868</p> | ||
| + | |||
| + | |||
| + | |||
| + | <p style="text-align: right"><i>Author: Andreas Hadjicharalambous and Sam Garforth</i></p> | ||
Latest revision as of 21:11, 24 October 2016
pCusC CusR RFP
Description
This synthetic promoter system includes the response regulator CusR (part of the CusR/S copper sensitive two-component system found in E. coli) and the an RFP variant (mKate2) expressed downstream of a copper sensitive promoter (pCusC). As CusR is found downstream of the promoter it activates means that a positive feedback loop is created when sufficient copper is sensed in the environment.
However we were unable to obtain of copy of this system without a point mutation which appears to have broken the feedback loop.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 527
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Our project was to investigate a probiotic treatment for the copper-accumulation disorder: Wilson's disease. This required a system able to detect dietary copper, ideally in the range over which copper concentration changes after a meal (around 5-10μM).
E. coli responds to copper is the CusS/CusR two-component system. This consists of the transmembrane histidine kinase enzyme: CusS in the bacterial cytoplasmic membrane and a cytoplasmic response regulator: CusR.
When CusS binds periplasmic copper it transfers a phosphate group from ATP to CusR aspartate residue 51 via CusS histidine residue 271. Phosphorylated CusR can bind to DNA inverted repeat CusR boxes (AAAATGACAANNTTGTCATTTT) and activate gene expression.
In E. coli this box is present between the promoters for CusCFBA operon which encodes a multi-protein pump that exports cytoplasmic and periplasmic copper from the cell and the CusRS operon which encodes the two component system (therefore acting as a positive feedback loop in vivo).(1)
A similar feedback loop with CusR was shown to be more sensitive to copper by Ravikumar et al.(2)
The following picture is a representation of how the construct works, theoretically in vivo:
Experience
We cloned pCusC CusR RFP from a gBlock into the shipping plasmid pSB1C3. E. coli strain MG1655 was transformed using the specific recombinant plasmid and a 5ml culture of a transformed colony was grown overnight. Flow cytometry and plate reader experiments were performed to test sensitivity of the construct to various copper concentration
Plate Reader
We tested the pCusC promoter with the fluorescent protein mKate2 (a form of RFP excitation/emission maxima at 588 and 633 nm).
To account for the number of cells present at different copper concentrations and different times we measured the optical density (OD) as a proportional measure of the number of cells present. For our parts using RFP we measured the optical density at 700nm for these parts as measuring OD600 would get interference from the fluorescent protein.
A range of copper concentrations were prepared from stock solutions. A large volume plate was then prepared with 10μl of copper solution, 10μl of overnight culture and 980μl of broth with antibiotic. This resulted in a 1 in 100 dilution of the copper solutions prepared.
This large-volume plate was then centrifuged to mix the solutions and then 200μl transferred to a small-volume plate with a clear lid and then placed in the plate reader.
Four biological repeats of the part to be tested in MG1655 at ten different copper concentrations were measured as well as four repeats of a MG1655 negative control.
The plate reader measured the fluorescence and OD every ten minutes for at least 12 hours, shaking between measurements.
Flow cytometry
Flow cytometry is a technique whereby cells are passed individually into the path of a beam of light. The frequency range of incident light can be adjusted to allow excitation of specific fluorophores in the sample of cells. Downstream detectors can measure fluorescent emission, and this data can be used to quantify the amount of fluorophore in each cell.
To ensure comparability of experiments, all cells were grown for 3-4hrs (until entrance into the exponential growth phase) at 37°C and 225rpm shaking in the presence of the inducer before measurement. This allowed adequate time for activation of expression by the promoter systems. The negative control used in all cases were MG1655 bacteria containing an empty shipping plasmid. As these did not contain any fluorescent molecules, this population could be used to set the negative “gate” (i.e. the background fluorescence of the bacterial cells). Although the experiments were tedious as every sample had to be measured manually, the results were of remarkably high quality, were clearly interpretable, and fit very well with the other experimental data.
Conclusions
This construct was an attempt to make a positive feedback system based upon pCusC. However, after many attempts we only managed to obtain a version with a point mutation (Val to Ala) in the CusR DNA-binding domain. The tests indicate that the mutation was tolerable but found no evidence of a copper sensitive system.
References
(1) Yamamoto K, Ishihama A. (2005) “Transcriptional response of Escherichia coli to external copper.” Mol Microbiol. 2005 Apr;56(1):215-27.
(2) Sambandam Ravikumar, Van Dung Pham, Seung Hwan Lee, Ik-keun Yoo, Soon Ho Hong (2012) “Modification of CusSR bacterial two-component systems by the introduction of an inducible positive feedback loop” Journal of Industrial Microbiology & Biotechnology June 2012, Volume 39, Issue 6, pp 861–868
Author: Andreas Hadjicharalambous and Sam Garforth