Difference between revisions of "Part:BBa K1998006"
SWinchester (Talk | contribs) |
Tom Collier (Talk | contribs) (→Part Verification) |
||
| (11 intermediate revisions by 3 users not shown) | |||
| Line 14: | Line 14: | ||
===Overview=== | ===Overview=== | ||
| − | This part is composed of the psbO, psbP, psbQ and psbR genes. The | + | This part is composed of the <i>psbO, psbP, psbQ</i> and <i>psbR</i> genes making up one of the operons in our PSII pathway. The first gene, <i>psbO</i> encodes a protein involved in stability. <i>psbP</i> gene optimises the availability of Ca2+ and Cl- cofactors in the Oxygen Evolving Complex in the PSII. The <i>psbQ</i> protein is an oxygen enhancer protein whilst the last gene, <i>psbR</i> is an important link the complex core involved in the assembly of the Oxygen Evolving Complex. |
| − | <br> | + | <br><br> |
These parts make up one of the operons in our PSII pathway. | These parts make up one of the operons in our PSII pathway. | ||
<br><br> | <br><br> | ||
| Line 21: | Line 21: | ||
===Biology & Literature=== | ===Biology & Literature=== | ||
| − | + | The gene, <i>psbO</i> in this operon is attached to the luminal membrane of PSII [1] and through a cluster of Mn2+ forms the centre of the oxygen evolving complex [2]. Described as having an eight strand β-barrel, a large loop between strands five and six connects the oxygen evolving complex to the luminal surface [3]. Inactivation of the <i>psbO</i> gene in Chlamydomonas reinhardtii prevents the assembly of PSII [4]. In Synechocystis sp. however, deletion of the <i>psbO</i> gene has not had a major effect on oxygen evolution, but a larger effect on the susceptibility of photo inhibition can be seen [5]. Therefore it can be concluded that the whilst the <i>psbO</i> gene is not essential to the assembly of the PSII complex or the water-splitting pathway it is important in providing protection to the PSII from damage caused by light. | |
| − | The psbP gene that comprises this part transcribes a protein that optimizes the availability of Ca2+ and Cl- cofactors in the Oxygen Evolving Complex in PSII to maintain the active Manganese cluster. | + | The <i>psbP</i> gene is one of the extrinsic proteins found in higher plants and algae [6]. It has a two domain structure made up of an antiparallel b-sheet and a b-strand that form domain 1. Domain 1 lies back to back with a central b-sheet (domain II) [7]. Decrease oxygen evolving activity was detected when <i>psbP</i> has not been present in C. reinhardtii [4]. The 23kDa protein is involved in PSII water oxidation process [8]. The <i>psbP</i> gene that comprises this part transcribes a protein that optimizes the availability of Ca2+ and Cl- cofactors in the Oxygen Evolving Complex in PSII to maintain the active Manganese cluster [9]. |
| − | The psbQ gene | + | The <i>psbQ</i> gene in C. reinhardtii binds directly the PSII complex independent of other extrinsic proteins [10]. The protein is found on the inner side of the thylakoid lumen with a polyproline II helix [11]. It helps aid PSII to function in conditions where there is low light [12]. Further to this, absence of <i>psbQ</i> has indicated an impairment of the ability of cells to generate oxygen as well as impacting on the presence of <i>psbV</i> destabilised [13]. |
| − | The psbR gene encodes a | + | The final gene in this operon is the <i>psbR</i>. This gene encodes a 10kDa protein and whilst the genes role is not certain, it is understood that the gene is necessary for the optimisation of electron transfer and water oxidation [14] as the absence of <i>psbR</i> causes instability in the PSII complex. Other <i>psbR</i> mutants have indicated similar decrease in oxygen evolution [15, 16]. |
| + | |||
| + | ===Part Verification=== | ||
| + | |||
| + | <html><centre><img src=" https://static.igem.org/mediawiki/2016/8/82/T--Macquarie_Australia--PSII_Show_Gel_and_.JPG " height="20%" width="40%"></center></html> | ||
| + | <br><br> | ||
| + | <b>Fig 1.</b> Gel electrophoresis of the operons and single parts constituting the Photosystem II pathway implemented in this project. The part psbOPQR 2588bp can be seen in lane 5 on the gel at the correct size relative to the 1kb ladder and other parts in our project shown on this gel. The band lower down in this lane on the gel is the pSB1C3 backbone 2070 bp. | ||
===Protein information=== | ===Protein information=== | ||
| − | psbO | + | <i>psbO</i> |
<br> | <br> | ||
mass: 27.96kDa | mass: 27.96kDa | ||
| Line 37: | Line 43: | ||
MAQKVGQAAAAAALATAMVAGSANALTFDEIQGLTYLQVKGSGIANTCPVLESGTTNLKELKAGSYKLENFC IEPTSFTVKEESQFKGGETEFVKTKLMTRLTYTLDAMSGSFKVGSDGSAELKEDDGIDYAATTVQLPGGERV AFLFTIKQFDGKGTLDNIKGDFLVPSYRGSSFLDPKGRGGSTGYDNAVALPARADAEELLKENVKITKALKG | MAQKVGQAAAAAALATAMVAGSANALTFDEIQGLTYLQVKGSGIANTCPVLESGTTNLKELKAGSYKLENFC IEPTSFTVKEESQFKGGETEFVKTKLMTRLTYTLDAMSGSFKVGSDGSAELKEDDGIDYAATTVQLPGGERV AFLFTIKQFDGKGTLDNIKGDFLVPSYRGSSFLDPKGRGGSTGYDNAVALPARADAEELLKENVKITKALKG | ||
SAVFSVAKVDPVTGEIAGVFESIQPSDTDLGAKPPKDIKVTGLWYAQLK*<br> | SAVFSVAKVDPVTGEIAGVFESIQPSDTDLGAKPPKDIKVTGLWYAQLK*<br> | ||
| − | + | <br> | |
| − | psbP<br> | + | <i>psbP</i><br> |
mass: 23.17kDa<br> | mass: 23.17kDa<br> | ||
sequence:<br> | sequence:<br> | ||
| Line 45: | Line 51: | ||
<br><br> | <br><br> | ||
| − | psbQ<br> | + | <i>psbQ</i><br> |
mass: 19.6kDa<br> | mass: 19.6kDa<br> | ||
sequence: <br> | sequence: <br> | ||
| − | MASGESRRAVLGGLLASAVAAVAPKAALALTPVDLFDDRSVRDRGFDLIYEARDLDLPQNVREGFTQARASL DETKKRVKESEARIDADLDVFIQKSYWTEAREQLRRQVGTLRFDLNTLASTKEKEAKKAALGLRKEFIQAVED | + | MASGESRRAVLGGLLASAVAAVAPKAALALTPVDLFDDRSVRDRGFDLIYEARDLDLPQNVREGFTQARASL |
| − | + | DETKKRVKESEARIDADLDVFIQKSYWTEAREQLRRQVGTLRFDLNTLASTKEKEAKKAALGLRKEFIQAVED | |
| − | <br><br> | + | <br>LDFALREKDQASAAKKLEITKAKLDSVLAAVL<br> |
| − | + | <br> | |
| − | psbR<br> | + | <i>psbR</i><br> |
mass: 12.2kDa<br> | mass: 12.2kDa<br> | ||
sequence:<br> | sequence:<br> | ||
| Line 59: | Line 65: | ||
===References=== | ===References=== | ||
| + | [1] De Las Rivas J, Barber J. Analysis of the structure of the <i>PsbO</i> protein and its implications. Photosynthesis research. 2004 Sep 1;81(3):329-43. | ||
| + | <br><br> | ||
| + | [2] Murata N, Miyao M. Extrinsic membrane proteins in the photosynthetic oxygen-evolving complex. Trends in Biochemical Sciences. 1985 Mar 1;10(3):122-4. | ||
| + | <br><br> | ||
| + | [3] Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004 Mar 19;303(5665):1831-8. | ||
| + | <br><br> | ||
| + | [4] Mayfield SP, Rahire M, Frank G, Zuber H, Rochaix JD. Expression of the nuclear gene encoding oxygen-evolving enhancer protein 2 is required for high levels of photosynthetic oxygen evolution in Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences. 1987 Feb 1;84(3):749-53. | ||
| + | <br><br> | ||
| + | [5] Mayes SR, Cook KM, Self SJ, Zhang Z, Barber J. Deletion of the gene encoding the Photosystem II 33 kDa protein from Synechocystis sp. PCC 6803 does not inactivate water-splitting but increases vulnerability to photoinhibition. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 1991 Sep 27;1060(1):1-2. | ||
| + | <br><br> | ||
| + | [6] Ifuku K, Yamamoto Y, Ono TA, Ishihara S, Sato F. PsbP protein, but not <i>PsbQ</i> protein, is essential for the regulation and stabilization of photosystem II in higher plants. Plant Physiology. 2005 Nov 1;139(3):1175-84. | ||
| + | <br><br> | ||
| + | [7] Kuwabara T, Suzuki K. Reversible changes in conformation of the 23-kDa protein of photosystem II and their relationship to the susceptibility of the protein to a proteinase from photosystem II membranes. Plant and cell physiology. 1995 Apr 1;36(3):495-504. | ||
| + | <br><br> | ||
| + | [8] Rova M, Franzén LG, Fredriksson PO, Styring S. Photosystem II in a mutant of Chlamydomonas reinhardtii lacking the 23 kDa <i>psbP</i> protein shows increased sensitivity to photoinhibition in the absence of chloride. Photosynthesis research. 1994 Jan 1;39(1):75-83. | ||
| + | <br><br> | ||
| + | [9] Ido K, Ifuku K, Yamamoto Y, Ishihara S, Murakami A, Takabe K, Miyake C, Sato F. Knockdown of the <i>psbP</i> protein does not prevent assembly of the dimeric PSII core complex but impairs accumulation of photosystem II supercomplexes in tobacco. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 2009 Jul 31;1787(7):873-81. | ||
| + | <br><br> | ||
| + | [10] Suzuki T, Minagawa J, Tomo T, Sonoike K, Ohta H, Enami I. Binding and functional properties of the extrinsic proteins in oxygen-evolving photosystem II particle from a green alga, Chlamydomonas reinhardtii having his-tagged CP47. Plant and cell physiology. 2003 Jan 15;44(1):76-84. | ||
| + | <br><br> | ||
| + | [11] Balsera M, Arellano JB, Revuelta JL, De las Rivas J, Hermoso JA. The 1.49 Å resolution crystal structure of <i>psbQ</i> from photosystem II of Spinacia oleracea reveals a PPII structure in the N-terminal region. Journal of molecular biology. 2005 Jul 29;350(5):1051-60. | ||
| + | <br><br> | ||
| + | [12] Yi X, Hargett SR, Frankel LK, Bricker TM. The <i>psbQ</i> protein is required in Arabidopsis for photosystem II assembly/stability and photoautotrophy under low light conditions. Journal of Biological Chemistry. 2006 Sep 8;281(36):26260-7. | ||
| + | <br><br> | ||
| + | [13] Kashino Y, Inoue-Kashino N, Roose JL, Pakrasi HB. Absence of the <i>psbQ</i> protein results in destabilization of the <i>psbV</i> protein and decreased oxygen evolution activity in cyanobacterial photosystem II. Journal of Biological Chemistry. 2006 Jul 28;281(30):20834-41. | ||
| + | <br><br> | ||
| + | [14] Vinyard DJ, Ananyev GM, Charles Dismukes G. Photosystem II: the reaction center of oxygenic photosynthesis*. Annual review of biochemistry. 2013 Jun 2;82:577-606. | ||
| + | <br><br> | ||
| + | [15] Stockhaus J, Höfer M, Renger G, Westhoff P, Wydrzynski T, Willmitzer L. Anti-sense RNA efficiently inhibits formation of the 10 kd polypeptide of photosystem II in transgenic potato plants: analysis of the role of the 10 kd protein. The EMBO journal. 1990 Sep;9(9):3013. | ||
| + | <br><br> | ||
| + | [16] Suorsa M, Sirpiö S, Allahverdiyeva Y, Paakkarinen V, Mamedov F, Styring S, Aro EM. PsbR, a missing link in the assembly of the oxygen-evolving complex of plant photosystem II. Journal of Biological Chemistry. 2006 Jan 6;281(1):145-50. | ||
Latest revision as of 03:41, 21 October 2016
psbOPQR
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1998
Illegal BglII site found at 2327 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 754
Illegal NgoMIV site found at 1400 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 2391
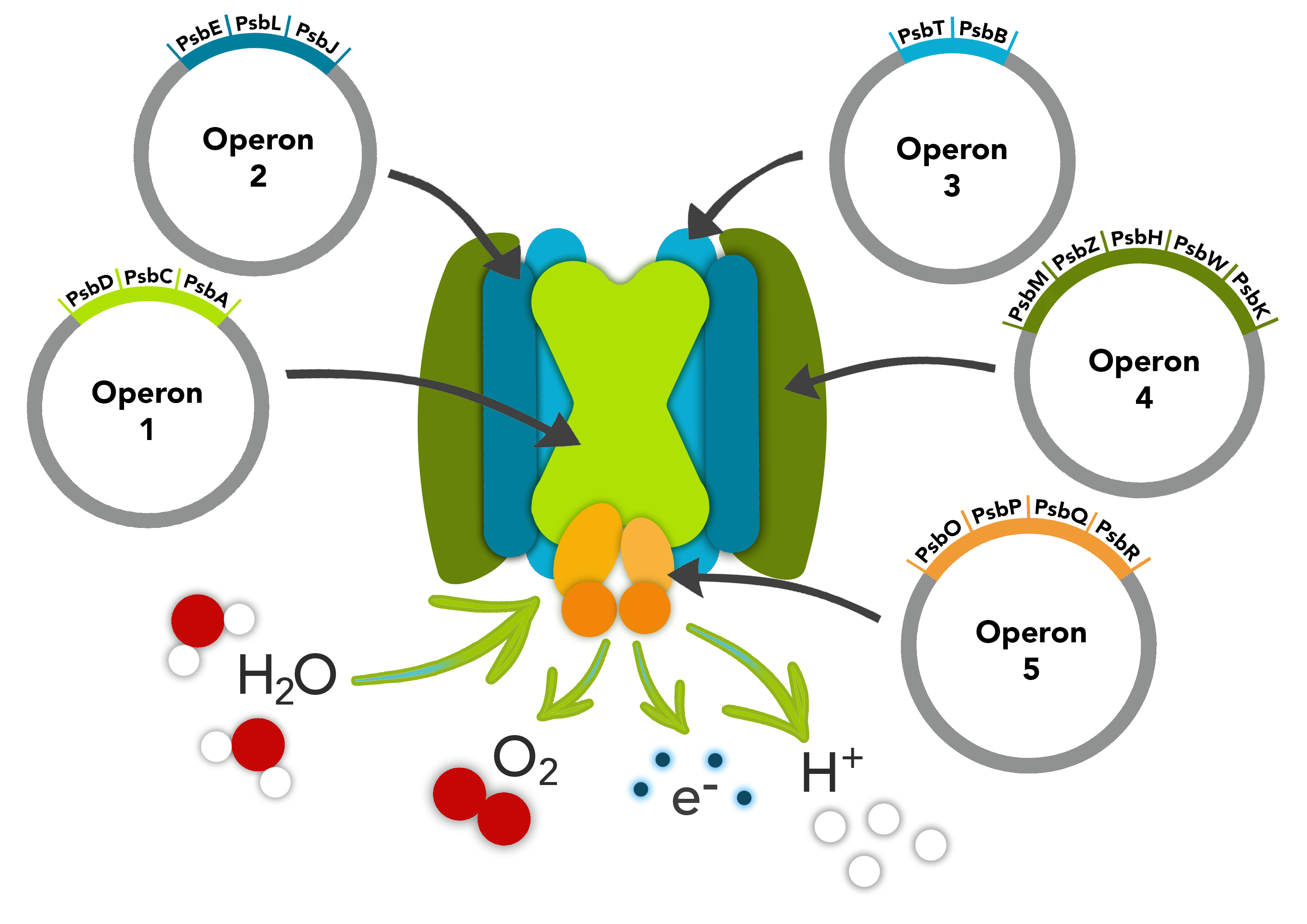
Overview
This part is composed of the psbO, psbP, psbQ and psbR genes making up one of the operons in our PSII pathway. The first gene, psbO encodes a protein involved in stability. psbP gene optimises the availability of Ca2+ and Cl- cofactors in the Oxygen Evolving Complex in the PSII. The psbQ protein is an oxygen enhancer protein whilst the last gene, psbR is an important link the complex core involved in the assembly of the Oxygen Evolving Complex.
These parts make up one of the operons in our PSII pathway.
Biology & Literature
The gene, psbO in this operon is attached to the luminal membrane of PSII [1] and through a cluster of Mn2+ forms the centre of the oxygen evolving complex [2]. Described as having an eight strand β-barrel, a large loop between strands five and six connects the oxygen evolving complex to the luminal surface [3]. Inactivation of the psbO gene in Chlamydomonas reinhardtii prevents the assembly of PSII [4]. In Synechocystis sp. however, deletion of the psbO gene has not had a major effect on oxygen evolution, but a larger effect on the susceptibility of photo inhibition can be seen [5]. Therefore it can be concluded that the whilst the psbO gene is not essential to the assembly of the PSII complex or the water-splitting pathway it is important in providing protection to the PSII from damage caused by light.
The psbP gene is one of the extrinsic proteins found in higher plants and algae [6]. It has a two domain structure made up of an antiparallel b-sheet and a b-strand that form domain 1. Domain 1 lies back to back with a central b-sheet (domain II) [7]. Decrease oxygen evolving activity was detected when psbP has not been present in C. reinhardtii [4]. The 23kDa protein is involved in PSII water oxidation process [8]. The psbP gene that comprises this part transcribes a protein that optimizes the availability of Ca2+ and Cl- cofactors in the Oxygen Evolving Complex in PSII to maintain the active Manganese cluster [9].
The psbQ gene in C. reinhardtii binds directly the PSII complex independent of other extrinsic proteins [10]. The protein is found on the inner side of the thylakoid lumen with a polyproline II helix [11]. It helps aid PSII to function in conditions where there is low light [12]. Further to this, absence of psbQ has indicated an impairment of the ability of cells to generate oxygen as well as impacting on the presence of psbV destabilised [13].
The final gene in this operon is the psbR. This gene encodes a 10kDa protein and whilst the genes role is not certain, it is understood that the gene is necessary for the optimisation of electron transfer and water oxidation [14] as the absence of psbR causes instability in the PSII complex. Other psbR mutants have indicated similar decrease in oxygen evolution [15, 16].
Part Verification

Fig 1. Gel electrophoresis of the operons and single parts constituting the Photosystem II pathway implemented in this project. The part psbOPQR 2588bp can be seen in lane 5 on the gel at the correct size relative to the 1kb ladder and other parts in our project shown on this gel. The band lower down in this lane on the gel is the pSB1C3 backbone 2070 bp.
Protein information
psbO
mass: 27.96kDa
sequence:
MAQKVGQAAAAAALATAMVAGSANALTFDEIQGLTYLQVKGSGIANTCPVLESGTTNLKELKAGSYKLENFC IEPTSFTVKEESQFKGGETEFVKTKLMTRLTYTLDAMSGSFKVGSDGSAELKEDDGIDYAATTVQLPGGERV AFLFTIKQFDGKGTLDNIKGDFLVPSYRGSSFLDPKGRGGSTGYDNAVALPARADAEELLKENVKITKALKG
SAVFSVAKVDPVTGEIAGVFESIQPSDTDLGAKPPKDIKVTGLWYAQLK*
psbP
mass: 23.17kDa
sequence:
MASGSDVSRRAALAGFAGAAALVSSSPANAAYGDSANVFGKVTNKSGFVPYAGDGFALLLPAKWNPSKENDF PGVILRYEDNFDAVNNLVVIAQDTDKKAIADFGSQDKFLESVSYLLGKQAYSGETQSEGGFAPNRVSAASLL DVSTTTDKKGKTYYKYELLVRSADGDEGGRHQLIGATVGSDNKLYIIKIQIGDKRWFKGAKKEAMGAFDSFTVV*
psbQ
mass: 19.6kDa
sequence:
MASGESRRAVLGGLLASAVAAVAPKAALALTPVDLFDDRSVRDRGFDLIYEARDLDLPQNVREGFTQARASL
DETKKRVKESEARIDADLDVFIQKSYWTEAREQLRRQVGTLRFDLNTLASTKEKEAKKAALGLRKEFIQAVED
LDFALREKDQASAAKKLEITKAKLDSVLAAVL
psbR
mass: 12.2kDa
sequence:
MGGGKTDITKVGLNSIEDPVVKQNLMGKSRFMNKKDWKDASGRKGKGYGVYRYEDKYGANVDGYSPIYTPDL
WTESGDSYTLGTKGLIAWAGLVLVLLAVGVNLIISTSQLGA
References
[1] De Las Rivas J, Barber J. Analysis of the structure of the PsbO protein and its implications. Photosynthesis research. 2004 Sep 1;81(3):329-43.
[2] Murata N, Miyao M. Extrinsic membrane proteins in the photosynthetic oxygen-evolving complex. Trends in Biochemical Sciences. 1985 Mar 1;10(3):122-4.
[3] Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004 Mar 19;303(5665):1831-8.
[4] Mayfield SP, Rahire M, Frank G, Zuber H, Rochaix JD. Expression of the nuclear gene encoding oxygen-evolving enhancer protein 2 is required for high levels of photosynthetic oxygen evolution in Chlamydomonas reinhardtii. Proceedings of the National Academy of Sciences. 1987 Feb 1;84(3):749-53.
[5] Mayes SR, Cook KM, Self SJ, Zhang Z, Barber J. Deletion of the gene encoding the Photosystem II 33 kDa protein from Synechocystis sp. PCC 6803 does not inactivate water-splitting but increases vulnerability to photoinhibition. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 1991 Sep 27;1060(1):1-2.
[6] Ifuku K, Yamamoto Y, Ono TA, Ishihara S, Sato F. PsbP protein, but not PsbQ protein, is essential for the regulation and stabilization of photosystem II in higher plants. Plant Physiology. 2005 Nov 1;139(3):1175-84.
[7] Kuwabara T, Suzuki K. Reversible changes in conformation of the 23-kDa protein of photosystem II and their relationship to the susceptibility of the protein to a proteinase from photosystem II membranes. Plant and cell physiology. 1995 Apr 1;36(3):495-504.
[8] Rova M, Franzén LG, Fredriksson PO, Styring S. Photosystem II in a mutant of Chlamydomonas reinhardtii lacking the 23 kDa psbP protein shows increased sensitivity to photoinhibition in the absence of chloride. Photosynthesis research. 1994 Jan 1;39(1):75-83.
[9] Ido K, Ifuku K, Yamamoto Y, Ishihara S, Murakami A, Takabe K, Miyake C, Sato F. Knockdown of the psbP protein does not prevent assembly of the dimeric PSII core complex but impairs accumulation of photosystem II supercomplexes in tobacco. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 2009 Jul 31;1787(7):873-81.
[10] Suzuki T, Minagawa J, Tomo T, Sonoike K, Ohta H, Enami I. Binding and functional properties of the extrinsic proteins in oxygen-evolving photosystem II particle from a green alga, Chlamydomonas reinhardtii having his-tagged CP47. Plant and cell physiology. 2003 Jan 15;44(1):76-84.
[11] Balsera M, Arellano JB, Revuelta JL, De las Rivas J, Hermoso JA. The 1.49 Å resolution crystal structure of psbQ from photosystem II of Spinacia oleracea reveals a PPII structure in the N-terminal region. Journal of molecular biology. 2005 Jul 29;350(5):1051-60.
[12] Yi X, Hargett SR, Frankel LK, Bricker TM. The psbQ protein is required in Arabidopsis for photosystem II assembly/stability and photoautotrophy under low light conditions. Journal of Biological Chemistry. 2006 Sep 8;281(36):26260-7.
[13] Kashino Y, Inoue-Kashino N, Roose JL, Pakrasi HB. Absence of the psbQ protein results in destabilization of the psbV protein and decreased oxygen evolution activity in cyanobacterial photosystem II. Journal of Biological Chemistry. 2006 Jul 28;281(30):20834-41.
[14] Vinyard DJ, Ananyev GM, Charles Dismukes G. Photosystem II: the reaction center of oxygenic photosynthesis*. Annual review of biochemistry. 2013 Jun 2;82:577-606.
[15] Stockhaus J, Höfer M, Renger G, Westhoff P, Wydrzynski T, Willmitzer L. Anti-sense RNA efficiently inhibits formation of the 10 kd polypeptide of photosystem II in transgenic potato plants: analysis of the role of the 10 kd protein. The EMBO journal. 1990 Sep;9(9):3013.
[16] Suorsa M, Sirpiö S, Allahverdiyeva Y, Paakkarinen V, Mamedov F, Styring S, Aro EM. PsbR, a missing link in the assembly of the oxygen-evolving complex of plant photosystem II. Journal of Biological Chemistry. 2006 Jan 6;281(1):145-50.
