Difference between revisions of "Part:BBa K1998002"
SWinchester (Talk | contribs) |
Tom Collier (Talk | contribs) (→Part Verification) |
||
| (7 intermediate revisions by 3 users not shown) | |||
| Line 18: | Line 18: | ||
===Overview=== | ===Overview=== | ||
| − | This biobrick contains three genes from the photosystem II complex of Chlamydomonas reinhardtii - psbT | + | This biobrick contains three genes from the photosystem II complex of Chlamydomonas reinhardtii - <i>psbT</i> and <i>psbB</i>. It is driven by a Plac promoter at the beginning of the sequence, with RBS’s preceding each gene, and terminator sequence at the 3' end. |
<br><br> | <br><br> | ||
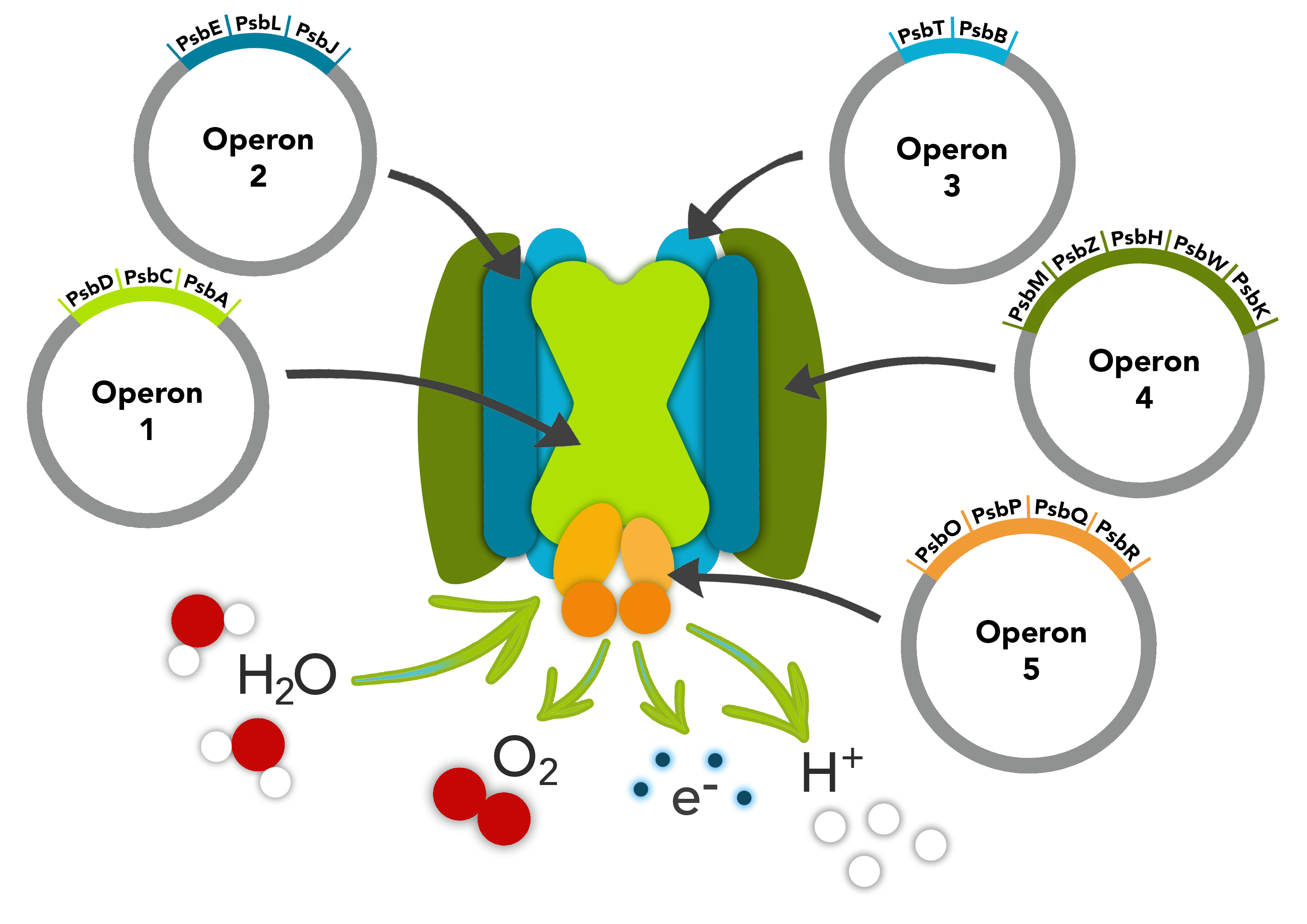
Parts composed in this operon:<br> | Parts composed in this operon:<br> | ||
| − | psbT - antenna proteins CP47 <br> | + | <i>psbT</i> - antenna proteins CP47 <br> |
| − | psbB - photosystem II (PSII) chlorophyll-binding protein<br> | + | <i>psbB</i> - photosystem II (PSII) chlorophyll-binding protein<br> |
These parts form operon 3 in our photosystem II pathway. | These parts form operon 3 in our photosystem II pathway. | ||
<br><br> | <br><br> | ||
| Line 28: | Line 28: | ||
===Biology & Literature=== | ===Biology & Literature=== | ||
| − | The second operon begins with psbT followed by psbB. Parts psbT and psbB make up the light harvesting complex inside the photosystem II complex [1]. They have a close interaction with the psbA and psbD complex helping to provide excitation energy from outer complexes onto them [2]. The psbT protein is a hydrophobic peptide that also has a single transmembrane helix which is associated with the PSII core complex [3] which binds to the complexes D1 and D2 heterodimer [4]. It's location in the core complex is believe to be at the PSII dimer interface [5]. Mutations in the psbT gene indicate that it's expression is not necessarily essential for the function of the PSII complex [4]. The role of psbT in repair of photo-damaged PSII complexes has also been discussed [3]. In addition the psbT gene maintains the stability of QA [5, 6]. | + | The second operon begins with <i>psbT</i> followed by <i>psbB</i>. Parts <i>psbT</i> and <i>psbB</i> make up the light harvesting complex inside the photosystem II complex [1]. They have a close interaction with the <i>psbA</i> and <i>psbD</i> complex helping to provide excitation energy from outer complexes onto them [2]. The <i>psbT</i> protein is a hydrophobic peptide that also has a single transmembrane helix which is associated with the PSII core complex [3] which binds to the complexes <i>D1</i> and <i>D2</i> heterodimer [4]. It's location in the core complex is believe to be at the PSII dimer interface [5]. Mutations in the <i>psbT</i> gene indicate that it's expression is not necessarily essential for the function of the PSII complex [4]. The role of <i>psbT</i> in repair of photo-damaged PSII complexes has also been discussed [3]. In addition the <i>psbT</i> gene maintains the stability of QA [5, 6]. |
<br><br> | <br><br> | ||
| − | The final gene of the composite part is psbB. The psbB protein is associated with the thylakoid membrane and forms part of the inner light harvesting complexes of PSII [7, 8]. | + | The final gene of the composite part is <i>psbB</i>. The <i>psbB</i> protein is associated with the thylakoid membrane and forms part of the inner light harvesting complexes of PSII [7, 8]. |
| + | |||
| + | ===Part Verification=== | ||
| + | |||
| + | <html><centre><img src=" https://static.igem.org/mediawiki/2016/8/82/T--Macquarie_Australia--PSII_Show_Gel_and_.JPG " height="20%" width="40%"></center></html> | ||
| + | <br><br> | ||
| + | <b>Fig 1.</b> Gel electrophoresis of the Photosystem II parts, here <i>psbTB</i> is shown to be the correct size of 1815bp (lane 3) with a cam backbone of 2,070bp, when compaired to 1kb ladder on the gel, which validates the Biobrick parts and backbones. | ||
===Protein information=== | ===Protein information=== | ||
| − | psbT<br> | + | <i>psbT</i><br> |
| − | mass: 3.6kDa | + | mass: 3.6kDa<br> |
| − | sequence: MEALVYTFLLVGTLGIIFFSIFFRDPPRMIK | + | sequence:<br> |
| + | MEALVYTFLLVGTLGIIFFSIFFRDPPRMIK | ||
<br><br> | <br><br> | ||
| − | psbB<br> | + | <i>psbB</i><br> |
| − | mass: 56.1kDa | + | mass: 56.1kDa<br> |
| − | sequence: | + | sequence: <br> |
| − | + | MGLPWYRVHTVVINDPGRLISVHLMHTALVSGWAGSMALFEISVFDPSDPVLNPMWRQGMFVLPFMTRLGITQSWGGWTISGETATNPGIWSYEGVAAAHIIL | |
| − | + | SGALFLASVWHWTYWDLELFRDPRTGKTALDLPKIFGIHLFLSGLLCFGFGAFHVTGVFGPGIWVSDPYGLTGRVQPVAPSWGADGFDPYNPGGIASHHIAAG | |
| − | + | ILGVLAGLFHLCVRPSIRLYFGLSMGSIETVLSSSIAAVFWAAFVVAGTMWYGSAATPIELFGPTRYQWDQGFFQQEIQKRVQASLAEGASLSDAWSRIPEKLAF | |
| − | + | YDYIGNNPAKGGLFRTGAMNSGDGIAVGWLGHASFKDQEGRELFVRRMPTFFETFPVLLLDKDGIVRADVPFRKAESKYSIEQVGVSVTFYGGELDGLTFTDP | |
| + | ATVKKYARKAQLGEIFEFDRSTLQSDGVFRSSPRGWFTFGHVCFALLFFFGHIWHGARTIFRDVFAGIDDDINDQVEFGKYKKLGDTSSLREAF | ||
<br> | <br> | ||
| Line 51: | Line 59: | ||
[2] Luciński R, Jackowski G. The structure, functions and degradation of pigment-binding proteins of photosystem II. Acta Biochim Pol. 2006;53(4):693-708. | [2] Luciński R, Jackowski G. The structure, functions and degradation of pigment-binding proteins of photosystem II. Acta Biochim Pol. 2006;53(4):693-708. | ||
<br><br> | <br><br> | ||
| − | [3] Monod C, Takahashi Y, Goldschmidt-Clermont M, Rochaix JD. The chloroplast ycf8 open reading frame encodes a photosystem II polypeptide which maintains photosynthetic activity under adverse growth conditions. The EMBO journal. 1994 Jun 15;13(12):2747. | + | [3] Monod C, Takahashi Y, Goldschmidt-Clermont M, Rochaix JD. The chloroplast <i>ycf8</i> open reading frame encodes a photosystem II polypeptide which maintains photosynthetic activity under adverse growth conditions. The EMBO journal. 1994 Jun 15;13(12):2747. |
<br><br> | <br><br> | ||
[4]Ohnishi N, Takahashi Y. PsbT polypeptide is required for efficient repair of photodamaged photosystem II reaction center. Journal of Biological Chemistry. 2001 Sep 7;276(36):33798-804. | [4]Ohnishi N, Takahashi Y. PsbT polypeptide is required for efficient repair of photodamaged photosystem II reaction center. Journal of Biological Chemistry. 2001 Sep 7;276(36):33798-804. | ||
| Line 57: | Line 65: | ||
[5] Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004 Mar 19;303(5665):1831-8. | [5] Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004 Mar 19;303(5665):1831-8. | ||
<br><br> | <br><br> | ||
| − | [6] Ohnishi N, Kashino Y, Satoh K, Ozawa SI, Takahashi Y. Chloroplast-encoded polypeptide | + | [6] Ohnishi N, Kashino Y, Satoh K, Ozawa SI, Takahashi Y. Chloroplast-encoded polypeptide <i>psbT</i> is involved in the repair of primary electron acceptor QA of photosystem II during photoinhibition in Chlamydomonas reinhardtii. Journal of Biological Chemistry. 2007 Mar 9;282(10):7107-15. |
<br><br> | <br><br> | ||
[7] Barber J, Nield J, Morris EP, Zheleva D, Hankamer B. The structure, function and dynamics of photosystem two. Physiologia plantarum. 1997 Aug 1;100(4):817-27. | [7] Barber J, Nield J, Morris EP, Zheleva D, Hankamer B. The structure, function and dynamics of photosystem two. Physiologia plantarum. 1997 Aug 1;100(4):817-27. | ||
<br><br> | <br><br> | ||
[8] Bricker TM, Frankel LK. The structure and function of CP47 and CP43 in photosystem II. Photosynthesis research. 2002 May 1;72(2):131-46. | [8] Bricker TM, Frankel LK. The structure and function of CP47 and CP43 in photosystem II. Photosynthesis research. 2002 May 1;72(2):131-46. | ||
Latest revision as of 03:40, 21 October 2016
psbTB
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Unknown
- 21INCOMPATIBLE WITH RFC[21]Unknown
- 23INCOMPATIBLE WITH RFC[23]Unknown
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Overview
This biobrick contains three genes from the photosystem II complex of Chlamydomonas reinhardtii - psbT and psbB. It is driven by a Plac promoter at the beginning of the sequence, with RBS’s preceding each gene, and terminator sequence at the 3' end.
Parts composed in this operon:
psbT - antenna proteins CP47
psbB - photosystem II (PSII) chlorophyll-binding protein
These parts form operon 3 in our photosystem II pathway.
Biology & Literature
The second operon begins with psbT followed by psbB. Parts psbT and psbB make up the light harvesting complex inside the photosystem II complex [1]. They have a close interaction with the psbA and psbD complex helping to provide excitation energy from outer complexes onto them [2]. The psbT protein is a hydrophobic peptide that also has a single transmembrane helix which is associated with the PSII core complex [3] which binds to the complexes D1 and D2 heterodimer [4]. It's location in the core complex is believe to be at the PSII dimer interface [5]. Mutations in the psbT gene indicate that it's expression is not necessarily essential for the function of the PSII complex [4]. The role of psbT in repair of photo-damaged PSII complexes has also been discussed [3]. In addition the psbT gene maintains the stability of QA [5, 6].
The final gene of the composite part is psbB. The psbB protein is associated with the thylakoid membrane and forms part of the inner light harvesting complexes of PSII [7, 8].
Part Verification

Fig 1. Gel electrophoresis of the Photosystem II parts, here psbTB is shown to be the correct size of 1815bp (lane 3) with a cam backbone of 2,070bp, when compaired to 1kb ladder on the gel, which validates the Biobrick parts and backbones.
Protein information
psbT
mass: 3.6kDa
sequence:
MEALVYTFLLVGTLGIIFFSIFFRDPPRMIK
psbB
mass: 56.1kDa
sequence:
MGLPWYRVHTVVINDPGRLISVHLMHTALVSGWAGSMALFEISVFDPSDPVLNPMWRQGMFVLPFMTRLGITQSWGGWTISGETATNPGIWSYEGVAAAHIIL
SGALFLASVWHWTYWDLELFRDPRTGKTALDLPKIFGIHLFLSGLLCFGFGAFHVTGVFGPGIWVSDPYGLTGRVQPVAPSWGADGFDPYNPGGIASHHIAAG
ILGVLAGLFHLCVRPSIRLYFGLSMGSIETVLSSSIAAVFWAAFVVAGTMWYGSAATPIELFGPTRYQWDQGFFQQEIQKRVQASLAEGASLSDAWSRIPEKLAF
YDYIGNNPAKGGLFRTGAMNSGDGIAVGWLGHASFKDQEGRELFVRRMPTFFETFPVLLLDKDGIVRADVPFRKAESKYSIEQVGVSVTFYGGELDGLTFTDP
ATVKKYARKAQLGEIFEFDRSTLQSDGVFRSSPRGWFTFGHVCFALLFFFGHIWHGARTIFRDVFAGIDDDINDQVEFGKYKKLGDTSSLREAF
References
[1] Barber J. Photosystem two. Biochimica et Biophysica Acta (BBA)-Bioenergetics. 1998 Jun 10;1365(1):269-77.
[2] Luciński R, Jackowski G. The structure, functions and degradation of pigment-binding proteins of photosystem II. Acta Biochim Pol. 2006;53(4):693-708.
[3] Monod C, Takahashi Y, Goldschmidt-Clermont M, Rochaix JD. The chloroplast ycf8 open reading frame encodes a photosystem II polypeptide which maintains photosynthetic activity under adverse growth conditions. The EMBO journal. 1994 Jun 15;13(12):2747.
[4]Ohnishi N, Takahashi Y. PsbT polypeptide is required for efficient repair of photodamaged photosystem II reaction center. Journal of Biological Chemistry. 2001 Sep 7;276(36):33798-804.
[5] Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004 Mar 19;303(5665):1831-8.
[6] Ohnishi N, Kashino Y, Satoh K, Ozawa SI, Takahashi Y. Chloroplast-encoded polypeptide psbT is involved in the repair of primary electron acceptor QA of photosystem II during photoinhibition in Chlamydomonas reinhardtii. Journal of Biological Chemistry. 2007 Mar 9;282(10):7107-15.
[7] Barber J, Nield J, Morris EP, Zheleva D, Hankamer B. The structure, function and dynamics of photosystem two. Physiologia plantarum. 1997 Aug 1;100(4):817-27.
[8] Bricker TM, Frankel LK. The structure and function of CP47 and CP43 in photosystem II. Photosynthesis research. 2002 May 1;72(2):131-46.
