Difference between revisions of "Part:BBa K1997005"
Zhuchushu13 (Talk | contribs) (→Functional Test) |
|||
| (9 intermediate revisions by 3 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K1997005 short</partinfo> | <partinfo>BBa_K1997005 short</partinfo> | ||
| − | This is | + | This part is an integrated tool for protein-protein interaction research using split-HRP system as reporter. the "FRB-RBS-FKBP" subpart can be easily replaced using Golden Gate technique with BsaI |
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | Intercellular protein–protein interactions (PPIs) enable communication between cells in diverse biological processes, including cell proliferation, immune responses, infection, and synaptic transmission, but they are challenging to visualize because existing techniques have insufficient sensitivity and/or specificity.<sup>1–3 </sup> | ||
| + | |||
| + | However, a split horseradish peroxidase (sHRP) has been reported as a sensitive and specific tool for the detection of intercellular PPIs. | ||
| + | |||
| + | The two sHRP fragments, engineered through screening of 17 cut sites in HRP followed by directed evolution, reconstitute into an active form when driven together by an intercellular PPI, producing bright fluorescence or contrast for electron microscopy. Fusing the sHRP fragments to the proteins neurexin (NRX) and neuroligin (NLG), which bind each other across the synaptic cleft<sup>4 </sup>, enabled sensitive visualization of synapses between specific sets of neurons, including two classes of synapses in the mouse visual system. sHRP should be widely applicable to studying mechanisms of communication between a variety of cell types. | ||
| + | |||
| + | ===Special Design=== | ||
| + | |||
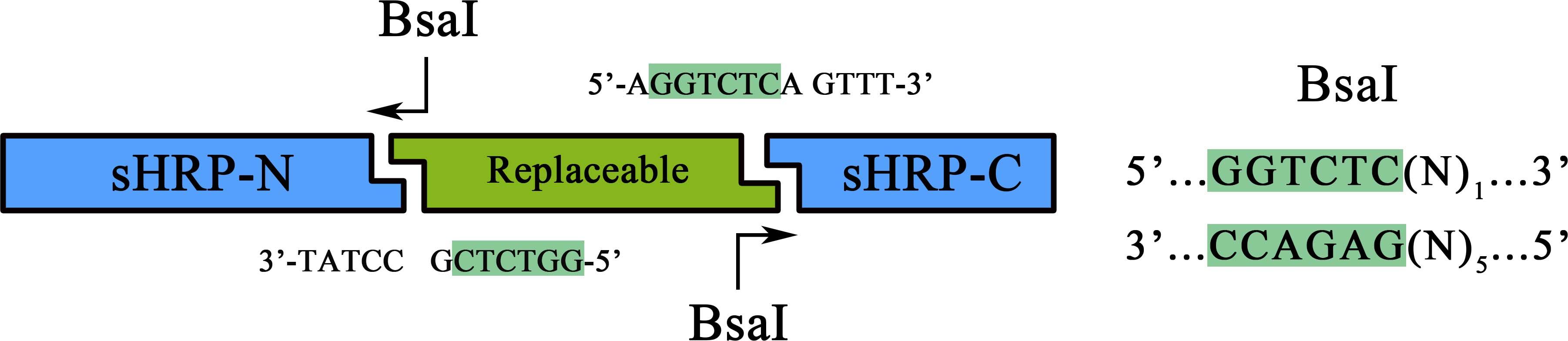
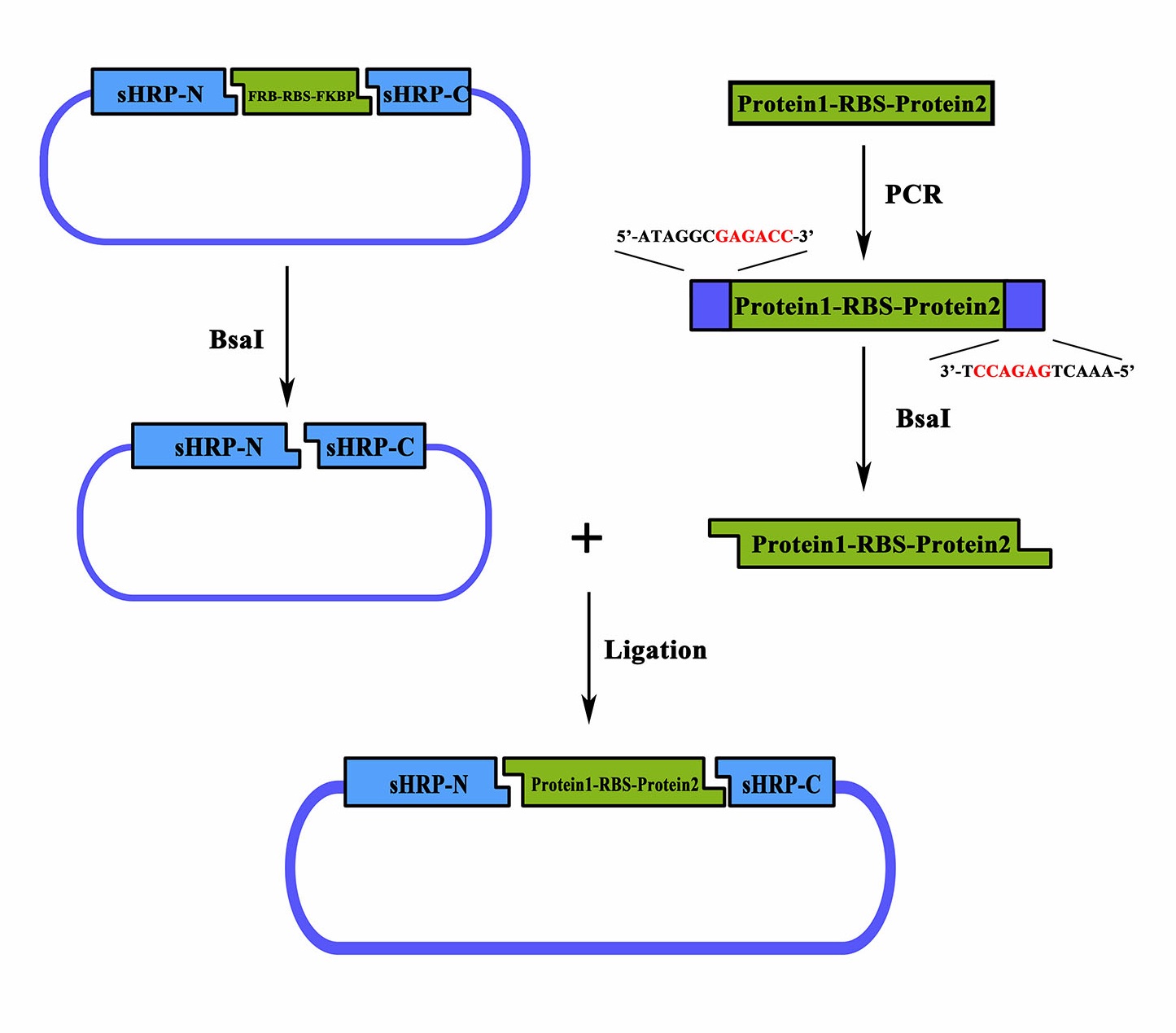
| + | As a member of the collection PPI tool kit, special designs were taken for to optimize the applicability and adaptive of such parts. Specifically, a novel designed substitution system, through which, two proteins could be fused with their corresponding split-HRP fragment at the same time using Golden-Gate Assembly, was introduced to dramatically simplify the cloning process). | ||
| + | |||
| + | [[File:sHRPfig1.jpg|500px|]] | ||
| + | |||
| + | [[File:NUDT-011-2.jpg|500px|]] | ||
| + | |||
| + | Figure 1. Schematic representation of the workflow of the substitution system | ||
| + | |||
| + | Coding sequence of proteins to be studied can be assembled with a RBS in between, a PCR procedure adding a 5’-ATAGGGGAGACC-3’ flank to the sense strand and a 3’-TCCAGAGTCAAA-5’ flank to the anti-sense would make it a proper substrate for the BsaI nuclease digest. Once digested, the product could be ligated together with the BsaI treated BBa_K1997005 to form a brand new device expressing the proteins of sHRP-N-Protein1, Protein2-sHRP-C and. The interaction between Protein1 and protein 2 could then be determined through the value of OD<sub>450 </sub> | ||
| + | |||
| + | |||
| + | ===Sequence and Features=== | ||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K1997005 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1997005 SequenceAndFeatures</partinfo> | ||
| + | ==Experimental Validation== | ||
| − | + | This part is validated through four ways: enzyme cutting, PCR, Sequence, and functional testing | |
| − | ===Functional | + | |
| − | < | + | ===Sequencing=== |
| − | + | ||
| + | This part is sequenced as correct after construction. | ||
| + | |||
| + | ===PCR=== | ||
| + | |||
| + | '''Methods''' | ||
| + | |||
| + | The PCR is performed with Premix EX Taq by Takara. | ||
| + | |||
| + | F-Prime: 5’- GAATTCGCGGCCGCTTCTAGAATGC-3’ | ||
| + | |||
| + | R-Prime: 5’- GGACTAGTATTATTGTTTGTCTGCC-3’ | ||
| + | |||
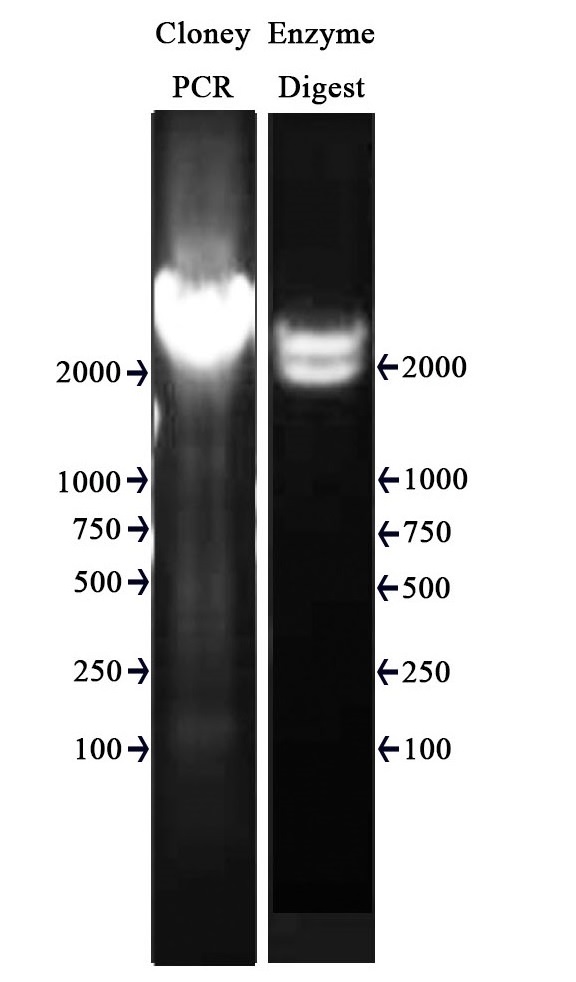
| + | The PCR protocol is selected based on the Users Manuel. | ||
| + | The Electrophoresis was performed on a 1% Agarose glu. | ||
| + | The result of the agarose electrophoresis was shown on the picture below. | ||
| + | |||
| + | [[File:NUDT-011-1.jpg|300px|]] | ||
| + | |||
| + | ===Enzyme digestion test === | ||
| + | |||
| + | '''Methods''' | ||
| + | |||
| + | After the assembly ,the plasmid was transferred into the Competent ''E. coli'' DH5α). After culturing overnight in LB,we minipreped the plasmid for cutting. | ||
| + | The preparation of the plasmid was performed with TIANprep Mini Plasmid Kit from ''TIANGEN''. The cutting procedure was performed with EcoRI and SpeI restriction endonuclease bought from ''TAKARA''. | ||
| + | |||
| + | The plasmid was cutted in a 20μL system at 37 ℃ for 2 hours. | ||
| + | The Electrophoresis was performed on a 1% Agarose glu. | ||
| + | |||
| + | The result of the agarose electrophoresis was shown on the picture above. | ||
| + | |||
| + | ===Functional Test=== | ||
| + | |||
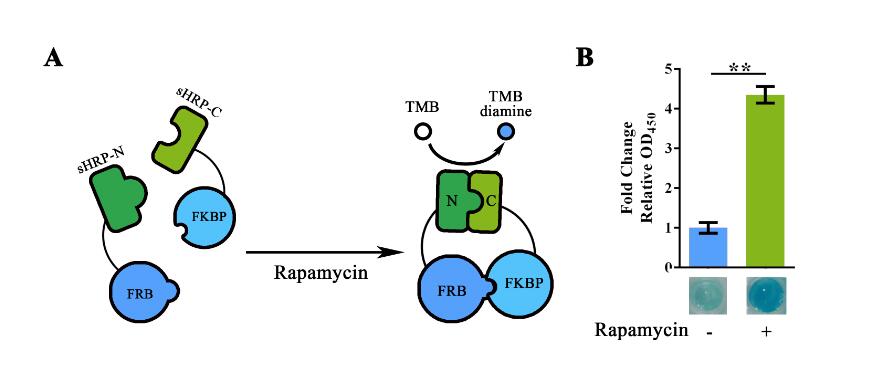
| + | The basic function of our split-HRP reporting system has been determined in the function validation of BBa_K1997013 and BBa_K1997012. Building on that, BBa_K1997005 was built to demonstrate the imbedded substitution system. For such matters, the Zif268 region in BBa_K1997012 was replaced into a “FRB-RBS-FKBP” fragment using Golden Gate Assembly. The cloning results were validated through sequencing. | ||
| + | |||
| + | For functional validation, we used Rapamycin to induce the interaction between FRB and FKBP. For such assay, E.coli carrying respective plasmid was cultured overnight under IPTG induction. Cells were then collected and lysed by high-pressure homogenizer. Once lysed and ultra-filtrated (to remove small molecules), 0.4nM of Rapamycin was added together with TMB substrate solution (with heme supplementation) into the cell lysate for the induction of protein-protein interaction and measurement of HRP activity. The plate was then incubated under 37°C for 5min before adding the Stop solution. | ||
| + | |||
| + | Value of OD<sub>450 </sub> obtained after adding the Stop Solution showed significant variation between the Rapamycin positive group and the Rapamycin negative group. Thus then validated the function of this part. | ||
| + | |||
| + | [[File:T--NUDT_CHINA--partsfig3.jpg|700px|]] | ||
| + | |||
| + | Figure 2. Rapamycin-induced sHRP-N-FRB/sHRP-C-FKBP interaction. (A) Schematic representation of the rapamycin induced protein-protein interaction. The adding of rapamycin would induce the interaction between FRB and FKBP, thus shortened the range between split-HRP fragments and reconstruct its structure. (B)Experiment showing the OD<sub>450 </sub> value with/without Rapamycin induction.. Relative OD<sub>450 </sub> value was calculated with normalization of the OD<sub>450 </sub> value. For Fold change Relative OD<sub>450 </sub> , relative OD<sub>450 </sub> of the group without Rapamycin induction was set arbitrarily as 1.0, and the levels of the other groups were adjusted correspondingly. The concentration of Rapamycin used in the experiment was 40nM. This experiment was run in three parallel reactions, and the data represent results obtained from at least three independent experiments. **p<0.01. | ||
| + | |||
| + | ===References=== | ||
| + | |||
| + | |||
| + | 1. Kim, S.A., Tai, C.-Y., Mok, L.-P., Mosser, E.A. & Schuman, E.M. Calcium-dependent | ||
| + | dynamics of cadherin interactions at cell-cell junctions. Proc. Natl. Acad. Sci. USA | ||
| + | 108, 9857–9862 (2011). | ||
| + | |||
| + | 2. Feinberg, E.H. et al. GFP Reconstitution Across Synaptic Partners (GRASP) defnes | ||
| + | cell contacts and synapses in living nervous systems. Neuron 57, 353–363 (2008). | ||
| + | |||
| + | 3. Liu, D.S., Loh, K.H., Lam, S.S., White, K.A. & Ting, A.Y. Imaging trans-cellular neurexinneuroligin interactions by enzymatic probe ligation. PLoS One 8, e52823 (2013). | ||
| + | |||
| + | 4. Craig, A.M. & Kang, Y. Neurexin-neuroligin signaling in synapse development. Curr. | ||
| + | Opin. Neurobiol. 17, 43–52 (2007). | ||
Latest revision as of 00:07, 21 October 2016
sHRP-N->FRB->RBS->FKBP-> sHRP-C
This part is an integrated tool for protein-protein interaction research using split-HRP system as reporter. the "FRB-RBS-FKBP" subpart can be easily replaced using Golden Gate technique with BsaI
Usage and Biology
Intercellular protein–protein interactions (PPIs) enable communication between cells in diverse biological processes, including cell proliferation, immune responses, infection, and synaptic transmission, but they are challenging to visualize because existing techniques have insufficient sensitivity and/or specificity.1–3
However, a split horseradish peroxidase (sHRP) has been reported as a sensitive and specific tool for the detection of intercellular PPIs.
The two sHRP fragments, engineered through screening of 17 cut sites in HRP followed by directed evolution, reconstitute into an active form when driven together by an intercellular PPI, producing bright fluorescence or contrast for electron microscopy. Fusing the sHRP fragments to the proteins neurexin (NRX) and neuroligin (NLG), which bind each other across the synaptic cleft4 , enabled sensitive visualization of synapses between specific sets of neurons, including two classes of synapses in the mouse visual system. sHRP should be widely applicable to studying mechanisms of communication between a variety of cell types.
Special Design
As a member of the collection PPI tool kit, special designs were taken for to optimize the applicability and adaptive of such parts. Specifically, a novel designed substitution system, through which, two proteins could be fused with their corresponding split-HRP fragment at the same time using Golden-Gate Assembly, was introduced to dramatically simplify the cloning process).
Figure 1. Schematic representation of the workflow of the substitution system
Coding sequence of proteins to be studied can be assembled with a RBS in between, a PCR procedure adding a 5’-ATAGGGGAGACC-3’ flank to the sense strand and a 3’-TCCAGAGTCAAA-5’ flank to the anti-sense would make it a proper substrate for the BsaI nuclease digest. Once digested, the product could be ligated together with the BsaI treated BBa_K1997005 to form a brand new device expressing the proteins of sHRP-N-Protein1, Protein2-sHRP-C and. The interaction between Protein1 and protein 2 could then be determined through the value of OD450
Sequence and Features
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 154
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 396
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1324
Illegal BsaI.rc site found at 679
Illegal SapI.rc site found at 1438
Experimental Validation
This part is validated through four ways: enzyme cutting, PCR, Sequence, and functional testing
Sequencing
This part is sequenced as correct after construction.
PCR
Methods
The PCR is performed with Premix EX Taq by Takara.
F-Prime: 5’- GAATTCGCGGCCGCTTCTAGAATGC-3’
R-Prime: 5’- GGACTAGTATTATTGTTTGTCTGCC-3’
The PCR protocol is selected based on the Users Manuel. The Electrophoresis was performed on a 1% Agarose glu. The result of the agarose electrophoresis was shown on the picture below.
Enzyme digestion test
Methods
After the assembly ,the plasmid was transferred into the Competent E. coli DH5α). After culturing overnight in LB,we minipreped the plasmid for cutting. The preparation of the plasmid was performed with TIANprep Mini Plasmid Kit from TIANGEN. The cutting procedure was performed with EcoRI and SpeI restriction endonuclease bought from TAKARA.
The plasmid was cutted in a 20μL system at 37 ℃ for 2 hours. The Electrophoresis was performed on a 1% Agarose glu.
The result of the agarose electrophoresis was shown on the picture above.
Functional Test
The basic function of our split-HRP reporting system has been determined in the function validation of BBa_K1997013 and BBa_K1997012. Building on that, BBa_K1997005 was built to demonstrate the imbedded substitution system. For such matters, the Zif268 region in BBa_K1997012 was replaced into a “FRB-RBS-FKBP” fragment using Golden Gate Assembly. The cloning results were validated through sequencing.
For functional validation, we used Rapamycin to induce the interaction between FRB and FKBP. For such assay, E.coli carrying respective plasmid was cultured overnight under IPTG induction. Cells were then collected and lysed by high-pressure homogenizer. Once lysed and ultra-filtrated (to remove small molecules), 0.4nM of Rapamycin was added together with TMB substrate solution (with heme supplementation) into the cell lysate for the induction of protein-protein interaction and measurement of HRP activity. The plate was then incubated under 37°C for 5min before adding the Stop solution.
Value of OD450 obtained after adding the Stop Solution showed significant variation between the Rapamycin positive group and the Rapamycin negative group. Thus then validated the function of this part.
Figure 2. Rapamycin-induced sHRP-N-FRB/sHRP-C-FKBP interaction. (A) Schematic representation of the rapamycin induced protein-protein interaction. The adding of rapamycin would induce the interaction between FRB and FKBP, thus shortened the range between split-HRP fragments and reconstruct its structure. (B)Experiment showing the OD450 value with/without Rapamycin induction.. Relative OD450 value was calculated with normalization of the OD450 value. For Fold change Relative OD450 , relative OD450 of the group without Rapamycin induction was set arbitrarily as 1.0, and the levels of the other groups were adjusted correspondingly. The concentration of Rapamycin used in the experiment was 40nM. This experiment was run in three parallel reactions, and the data represent results obtained from at least three independent experiments. **p<0.01.
References
1. Kim, S.A., Tai, C.-Y., Mok, L.-P., Mosser, E.A. & Schuman, E.M. Calcium-dependent dynamics of cadherin interactions at cell-cell junctions. Proc. Natl. Acad. Sci. USA 108, 9857–9862 (2011).
2. Feinberg, E.H. et al. GFP Reconstitution Across Synaptic Partners (GRASP) defnes cell contacts and synapses in living nervous systems. Neuron 57, 353–363 (2008).
3. Liu, D.S., Loh, K.H., Lam, S.S., White, K.A. & Ting, A.Y. Imaging trans-cellular neurexinneuroligin interactions by enzymatic probe ligation. PLoS One 8, e52823 (2013).
4. Craig, A.M. & Kang, Y. Neurexin-neuroligin signaling in synapse development. Curr. Opin. Neurobiol. 17, 43–52 (2007).