Difference between revisions of "Part:BBa K2120207"
| Line 4: | Line 4: | ||
Use RFP to indicate the strength of promoters. Ptet-25 is the mutation of Ptet.The difference is the -35 region. See details in sequence. | Use RFP to indicate the strength of promoters. Ptet-25 is the mutation of Ptet.The difference is the -35 region. See details in sequence. | ||
| + | |||
| + | ===A series of Ptet promoters=== | ||
| + | |||
| + | We designed a killer switch controlled by TetR repressor. When the concentration of inhibitors reduces to a threshold, the downstream in-promoter Ptet will start the transcription of killer gene. After measuring the original promoter strength [Part:BBa_R0040], we found that the activity is too high to be well controlled for expressing killer genes. So we mutated the original promoter and got 4 mutants with lower activity. | ||
| + | |||
| + | '''Design''' | ||
| + | |||
| + | We analyzed the promoter region and decided to mutated the -35 region through designing random primers. It can at most ensure the change of promoter’s background activity and maintain the promoter’s character since the tightly binding region with repressor is well-conserved while the tightly binding region with RNA polymerase is changed . | ||
| + | |||
| + | '''Characterization using RFP''' | ||
| + | |||
| + | The four mutated promoters were characterized using RFP to measure the level of gene expression. Cells were grown in 1.5 mL tube in LB culture medium. The host we used is E.coli TOP10. | ||
| + | |||
| + | '''Results''' | ||
| + | |||
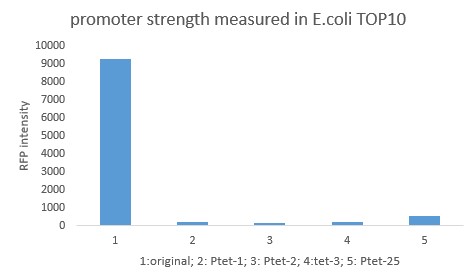
| + | Cells showed Red fluorescence under fluorescence microscopy. Quantitative measurement was carried out through enzyme-labeled instrument. It turned out that the highest activity of mutants is about 500, while the common activity is about 200. The original promoter is 9000. | ||
| + | [[File:BIT-CHINA-PARTS-MUTATION-1.jpg|600px|thumb|center|Fig1. Compared with the original promoter, the promoter strength of mutants Ptet-1, Ptet-2, Ptet-3, Ptet-25 is sharply decreased.]] | ||
| + | |||
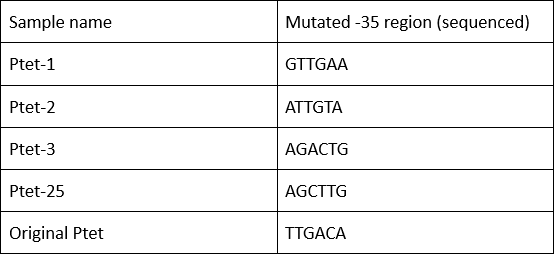
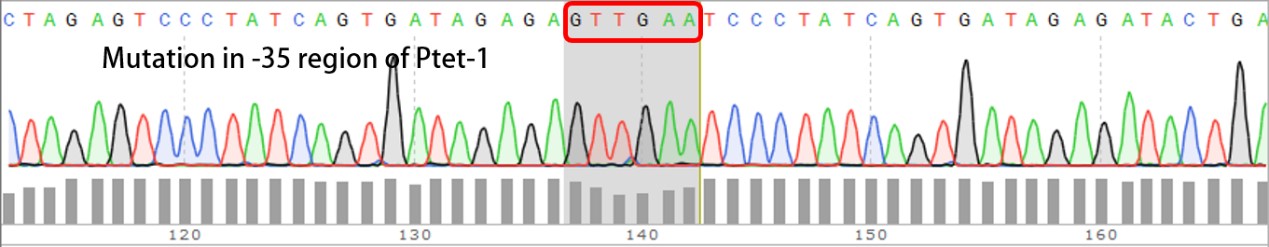
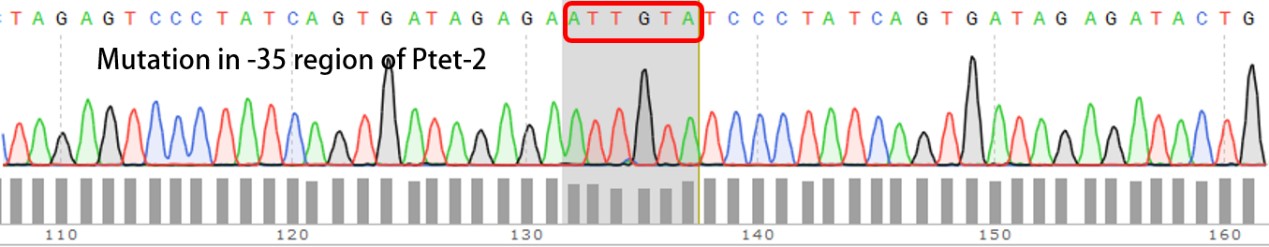
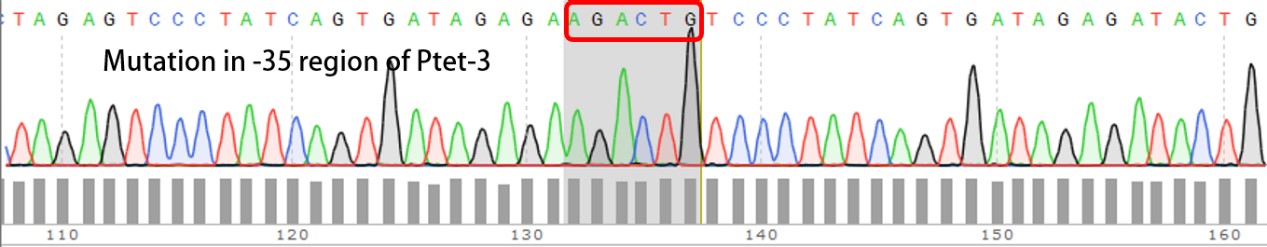
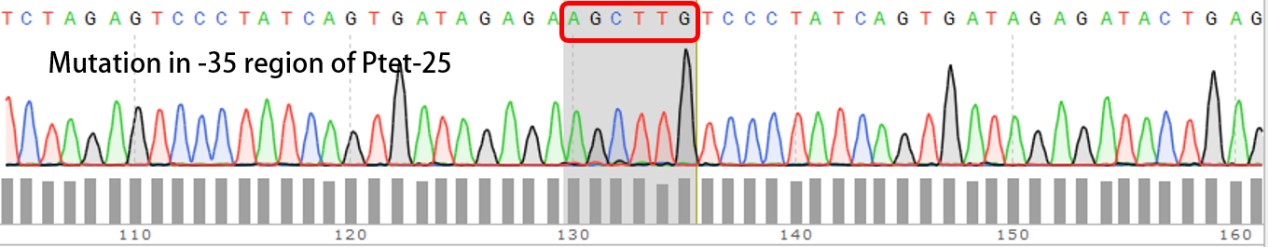
| + | We have submitted four mutated promoters [[https://parts.igem.org/wiki/index.php?title=Part:BBa_K2120200 BBa_K2120200]; [https://parts.igem.org/wiki/index.php?title=Part:BBa_K2120201 BBa_K2120201]; [https://parts.igem.org/wiki/index.php?title=Part:BBa_K2120202 BBa_K2120202] ; [https://parts.igem.org/wiki/index.php?title=Part:BBa_K2120203 BBa_K2120203]], here we listed the mutation in -35 region: | ||
| + | [[File:BIT-CHINA-PARTS-MUTATION-6.png|600px|thumb|center]] | ||
| + | [[File:BIT-CHINA-PARTS-MUTATION-2.jpg|600px|thumb|center]] | ||
| + | [[File:BIT-CHINA-PARTS-MUTATION-3.jpg|600px|thumb|center]] | ||
| + | [[File:BIT-CHINA-PARTS-MUTATION-4.jpg|600px|thumb|center]] | ||
| + | [[File:BIT-CHINA-PARTS-MUTATION-5.jpg|600px|thumb|center]] | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 16:30, 20 October 2016
Ptet-25+B0034+RFP+B0015
Use RFP to indicate the strength of promoters. Ptet-25 is the mutation of Ptet.The difference is the -35 region. See details in sequence.
A series of Ptet promoters
We designed a killer switch controlled by TetR repressor. When the concentration of inhibitors reduces to a threshold, the downstream in-promoter Ptet will start the transcription of killer gene. After measuring the original promoter strength [Part:BBa_R0040], we found that the activity is too high to be well controlled for expressing killer genes. So we mutated the original promoter and got 4 mutants with lower activity.
Design
We analyzed the promoter region and decided to mutated the -35 region through designing random primers. It can at most ensure the change of promoter’s background activity and maintain the promoter’s character since the tightly binding region with repressor is well-conserved while the tightly binding region with RNA polymerase is changed .
Characterization using RFP
The four mutated promoters were characterized using RFP to measure the level of gene expression. Cells were grown in 1.5 mL tube in LB culture medium. The host we used is E.coli TOP10.
Results
Cells showed Red fluorescence under fluorescence microscopy. Quantitative measurement was carried out through enzyme-labeled instrument. It turned out that the highest activity of mutants is about 500, while the common activity is about 200. The original promoter is 9000.
We have submitted four mutated promoters [BBa_K2120200; BBa_K2120201; BBa_K2120202 ; BBa_K2120203], here we listed the mutation in -35 region:
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 635
Illegal AgeI site found at 747 - 1000COMPATIBLE WITH RFC[1000]