Difference between revisions of "Part:BBa K1933100"
(→Western blotting) |
(→Fluorescence Microscopy) |
||
| (30 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1933100 short</partinfo> | <partinfo>BBa_K1933100 short</partinfo> | ||
| − | CBDcex fused to INPNC with 6xHis tag is one of a series of surface expressing fusion proteins that make up biodevice that aims to be therapeutic solution against | + | CBDcex fused to INPNC with 6xHis tag is one of a series of surface expressing fusion proteins that make up biodevice that aims to be the therapeutic solution against Norovirus infection. This protein in particular is a cellulose binding domain(CBDcex) fused to surface expression anchoring domain(INPNC), connected by a 6xHis tag to be easily identified by Western blotting. |
| + | <br>For more information, please visit ''' [http://2016.igem.org/Team:Kyoto our wiki] '''. | ||
==Sequence and Features== | ==Sequence and Features== | ||
| Line 10: | Line 10: | ||
==Usage== | ==Usage== | ||
| − | + | We succeeded in surface expressing anti-NoV scFv [https://parts.igem.org/Part:BBa_K1933002 BBa_K1933002] antibody using INPNC-His. We aimed to orally administrate this protein expressing '' E. coli, '' however, we obtained an overwhelmingly negative result from surveys conducted when asked "Would you feel uncomfortable with oral administration of sterilized recombinant organisms?" This is why we also constructed a parts that surface expresses cellulose binding domains (CBD). With this part, we can "leash" '' E.coli '' to cellulose and make them swiftly exit human digestive systems. We selected CBDcex as a CBD for this part. | |
| − | + | <br>This part had an important role in integrating our [http://2016.igem.org/Team:Kyoto/HP/Gold Human Practice] activities into our project. | |
| − | + | ||
==Characterization== | ==Characterization== | ||
| − | + | ''' Sequence confirmed! ''' | |
| − | + | ||
| − | + | ||
===RT-PCR=== | ===RT-PCR=== | ||
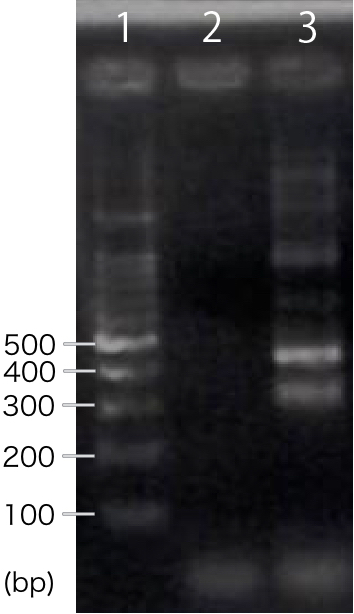
| − | We designed PCR primers so that we would obtain PCR product with the length of | + | We designed PCR primers so that we would obtain PCR product with the length of 311 bp after RT-PCR. For negative control, reverse transcription was not performed. |
[[file:T--Kyoto--RT-CBDcex.001.jpeg|200px|thumb|center|'''Fig.1''': ''' RT-PCR '''<br>1. marker, 2. INP-His-CBDcex (negative control), 3. INP-His-CBDcex]] | [[file:T--Kyoto--RT-CBDcex.001.jpeg|200px|thumb|center|'''Fig.1''': ''' RT-PCR '''<br>1. marker, 2. INP-His-CBDcex (negative control), 3. INP-His-CBDcex]] | ||
| − | A band corresponding to | + | A band corresponding to 311 bp was observed only in INP-His-scFv sample, not in negative control(Fig.1). This result suggests that INP-His-CBDcex was successfully transcribed. |
===Western blotting=== | ===Western blotting=== | ||
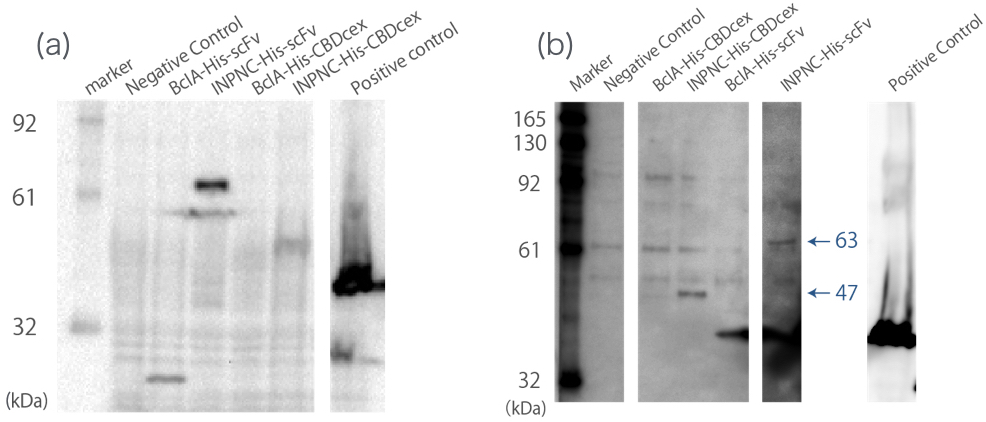
| − | [[file:T--Kyoto--Western blotting Whole cell and membrane fraction.jpeg|600px|thumb|center|'''Fig.2''': ''' (a) '''whole cell Western blotting against His tag. ''' (b) '''membrane fraction Western blotting against His tag.<br> Negative Control:BBa_K165002, BclA-His-scFv:33 kDa, INPNC-His-scFv:63 kDa, BclA-His-CBDcex:17 kDa, INPNC-His-CBDcex:47 kDa, Positive control | + | We used whole-cell Western blotting with anti-His tag antibody(Fig.2(a)). Then, we prepared the membrane fraction from the '' E. coli '' lysate. Membrane fraction was solubilized and used for Nickel Sepharose purification and precipitates were examined by Western blotting against His tag(Fig.2(b)). |
| − | A band corresponding to INPNC-His-CBDcex (47kDa) was observed in both whole cell and membrane fraction Western blotting (Fig.2(a),(b)), which confirms expression of the fusion proteins. | + | [[file:T--Kyoto--Western blotting Whole cell and membrane fraction.jpeg|600px|thumb|center|'''Fig.2''': ''' (a) '''whole cell Western blotting against His tag. ''' (b) '''membrane fraction Western blotting against His tag.<br> Negative Control:BBa_K165002, BclA-His-scFv:33 kDa, INPNC-His-scFv:63 kDa, BclA-His-CBDcex:17 kDa, INPNC-His-CBDcex:47 kDa, Positive control(Lpp-OmpA-scFv-His):43 kDa]] |
| + | A band corresponding to INPNC-His-CBDcex (47kDa) was observed in both whole cell and membrane fraction Western blotting (Fig.2(a),(b)), which confirms expression of the fusion proteins, and suggests that CBDcex was surface expressed, respectively. | ||
| − | == | + | ===Fluorescence Microscopy=== |
| + | As shown in Fig3, we observed the expression of INPNC-His-CBDcex on the membrane fraction. We next tested CBDcex's binding to cellulose. | ||
| + | [[file:T--Kyoto--projectfig16.png|400px|thumb|center|'''Fig3''': ''' Binding of '' E. coli '' to Cellulose with Fluorescence Microscopy '''<br>a-1. Bemliese (5mmx5mm) only, a-2. INPNC-His-ctl (OD600=0.2, 1ml) with Bemliese(5mmx5mm), a-3. INPNC-His-CBDcex (OD600=0.2, 1ml) with Bemliese (5mmx5mm) with INPNC-His-CBDcex (OD600=0.2, 1ml), b. cellulose powder (0.5mg) only, c-1. INPNC-His-CBDctl (OD600=1.0, 1ml) with cellulose powder (0.5mg), c-2. INPNC-His-CBDcex (OD600=1.0, 1ml) with cellulose powder (0.5mg), d-1. INPNC-His-CBDctl (OD600=1.0, 1ml) with cellulose powder (0.1mg), d-2 INPNC-His-CBDcex (OD600=1.0, 1ml) with cellulose powder (0.1mg)<br> | ||
| + | Fluorescent objects are the DAPI stained '' E. coli. '']] | ||
| + | [[file:T--Kyoto--projectfig17.png|600px|thumb|center|'''Fig4''':Quantification of binding efficiency of <i>E. coli</i> to cellulose<br> | ||
| + | Sample names correspond with Sample names of Fig16.<br> | ||
| + | (1) Binding of <i>E. coli</i> to bemliese<br> | ||
| + | The results of F test and T test between a-2 and a-3; F(10,12)=5.50 ,p<0.01, t(11)=2.28, p<0.05<br> | ||
| + | (2) Binding of <i>E. coli</i> to cellulose powder<br> | ||
| + | The results of F test and T test between c-1 and c-2; F(8,9)=33.88, p<0.01, t(8)=3.24, p<0.05<br> | ||
| + | The results of F test and T test between d-1 and d-2; F(10,10)=18.64, p<0.01, t(11)=3.44, p<0.01</div>]] | ||
| + | Fig3 shows the results. After incubating '' E. coli '' with about 35~38 micrometer cellulose powder, we DAPI stained '' E. coli '' and observed it under fluorescence microscopy. As shown, CBD-surface expressing '' E. coli '' was observed binding to cellulose, showing the ''' functional expression of INPNC-His-CBD ''' in our system. When control plasmid was used, the numbers of cellulose-bound <i>E. coli</i> cells were significantly reduced (Fig4). <br> | ||
| + | From these results, we concluded that this part can be used to physically link <i>E. coli</i> and cellulose. | ||
==''' Judging '''== | ==''' Judging '''== | ||
Latest revision as of 13:03, 20 October 2016
constitutive expression of CBDcex fused to INPNC with 6xHis tag
CBDcex fused to INPNC with 6xHis tag is one of a series of surface expressing fusion proteins that make up biodevice that aims to be the therapeutic solution against Norovirus infection. This protein in particular is a cellulose binding domain(CBDcex) fused to surface expression anchoring domain(INPNC), connected by a 6xHis tag to be easily identified by Western blotting.
For more information, please visit [http://2016.igem.org/Team:Kyoto our wiki] .
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 1012
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 132
Illegal NgoMIV site found at 465
Illegal AgeI site found at 883 - 1000COMPATIBLE WITH RFC[1000]
Usage
We succeeded in surface expressing anti-NoV scFv BBa_K1933002 antibody using INPNC-His. We aimed to orally administrate this protein expressing E. coli, however, we obtained an overwhelmingly negative result from surveys conducted when asked "Would you feel uncomfortable with oral administration of sterilized recombinant organisms?" This is why we also constructed a parts that surface expresses cellulose binding domains (CBD). With this part, we can "leash" E.coli to cellulose and make them swiftly exit human digestive systems. We selected CBDcex as a CBD for this part.
This part had an important role in integrating our [http://2016.igem.org/Team:Kyoto/HP/Gold Human Practice] activities into our project.
Characterization
Sequence confirmed!
RT-PCR
We designed PCR primers so that we would obtain PCR product with the length of 311 bp after RT-PCR. For negative control, reverse transcription was not performed.
A band corresponding to 311 bp was observed only in INP-His-scFv sample, not in negative control(Fig.1). This result suggests that INP-His-CBDcex was successfully transcribed.
Western blotting
We used whole-cell Western blotting with anti-His tag antibody(Fig.2(a)). Then, we prepared the membrane fraction from the E. coli lysate. Membrane fraction was solubilized and used for Nickel Sepharose purification and precipitates were examined by Western blotting against His tag(Fig.2(b)).
A band corresponding to INPNC-His-CBDcex (47kDa) was observed in both whole cell and membrane fraction Western blotting (Fig.2(a),(b)), which confirms expression of the fusion proteins, and suggests that CBDcex was surface expressed, respectively.
Fluorescence Microscopy
As shown in Fig3, we observed the expression of INPNC-His-CBDcex on the membrane fraction. We next tested CBDcex's binding to cellulose.
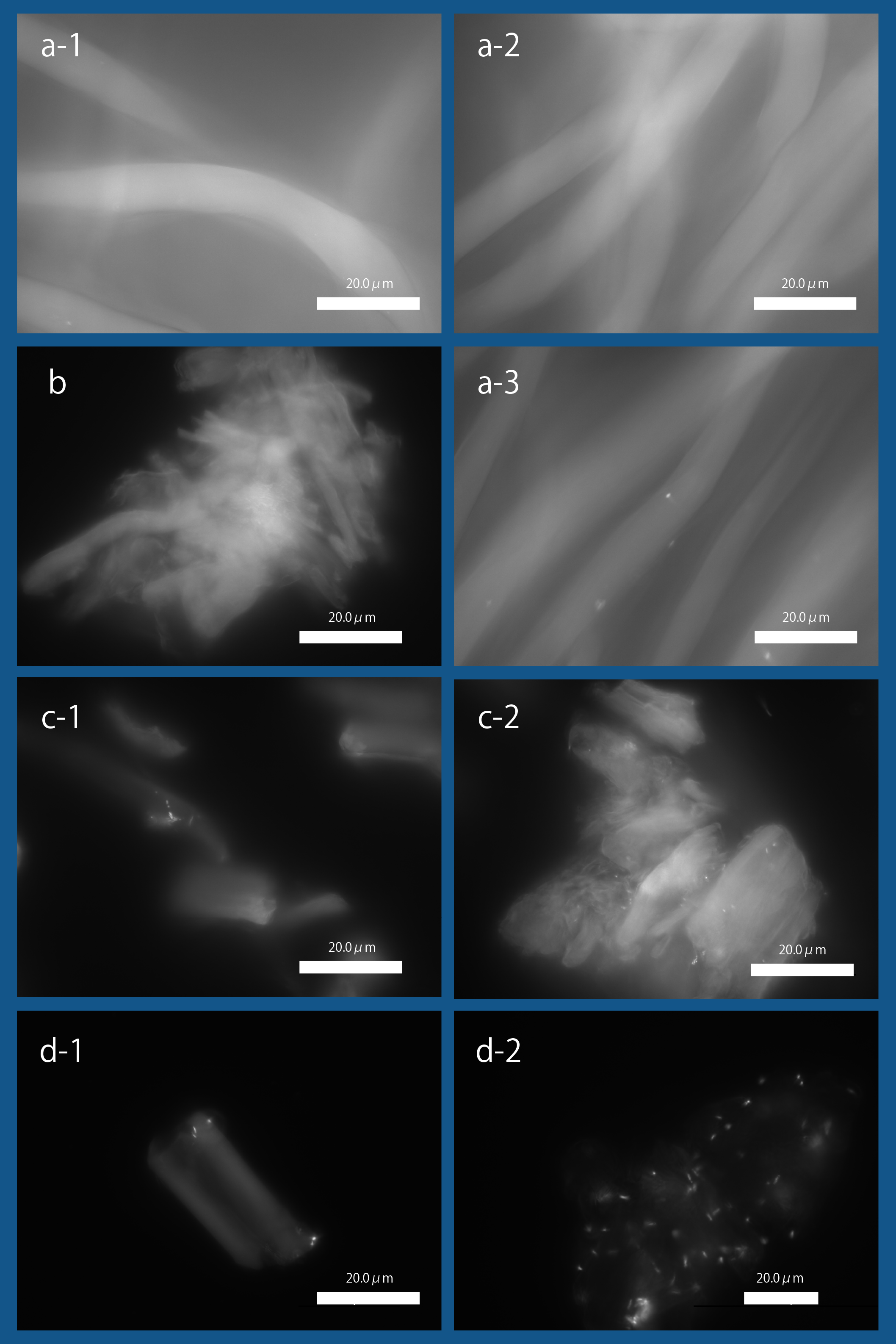
a-1. Bemliese (5mmx5mm) only, a-2. INPNC-His-ctl (OD600=0.2, 1ml) with Bemliese(5mmx5mm), a-3. INPNC-His-CBDcex (OD600=0.2, 1ml) with Bemliese (5mmx5mm) with INPNC-His-CBDcex (OD600=0.2, 1ml), b. cellulose powder (0.5mg) only, c-1. INPNC-His-CBDctl (OD600=1.0, 1ml) with cellulose powder (0.5mg), c-2. INPNC-His-CBDcex (OD600=1.0, 1ml) with cellulose powder (0.5mg), d-1. INPNC-His-CBDctl (OD600=1.0, 1ml) with cellulose powder (0.1mg), d-2 INPNC-His-CBDcex (OD600=1.0, 1ml) with cellulose powder (0.1mg)
Fluorescent objects are the DAPI stained E. coli.
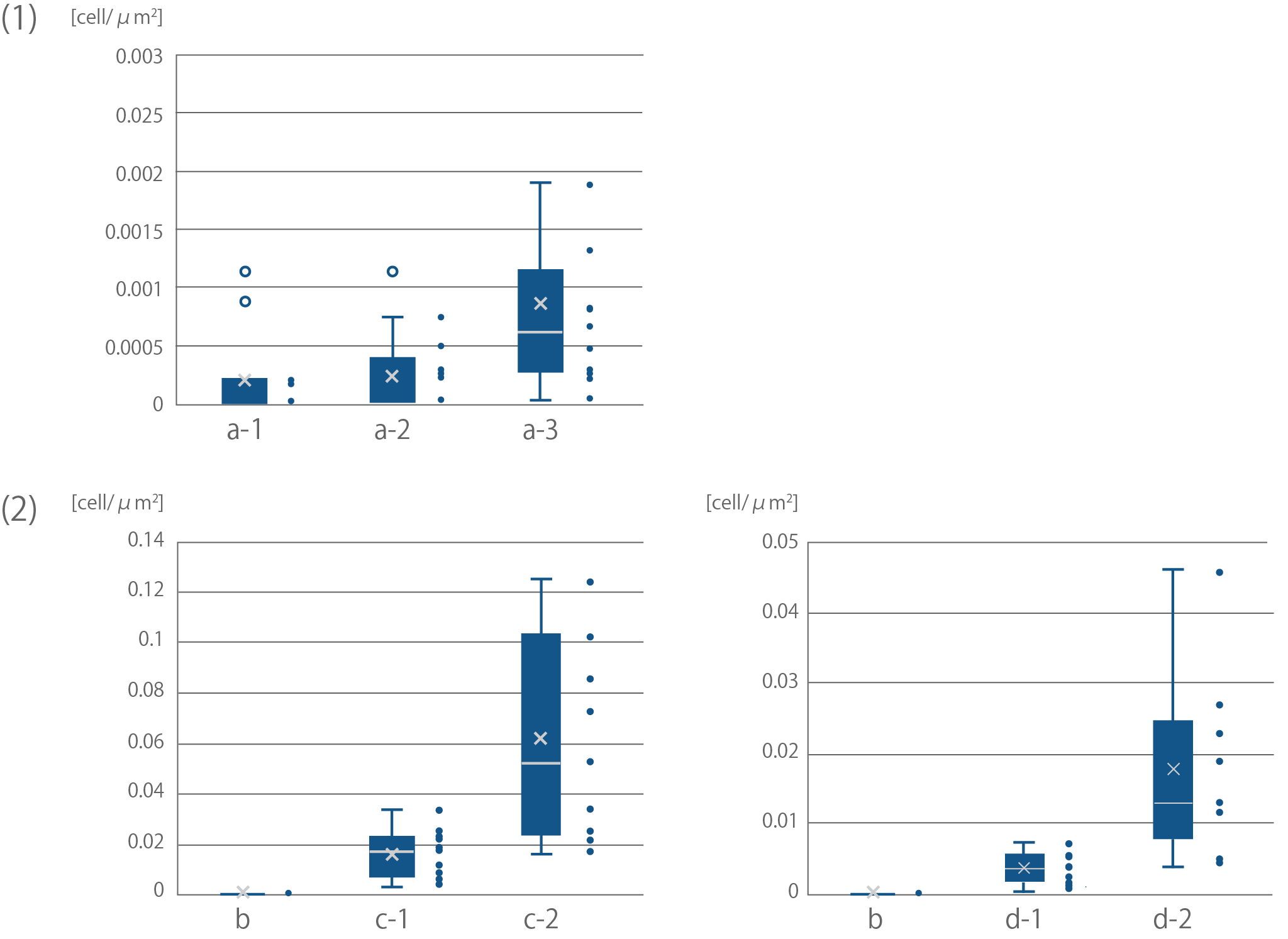
Sample names correspond with Sample names of Fig16.
(1) Binding of E. coli to bemliese
The results of F test and T test between a-2 and a-3; F(10,12)=5.50 ,p<0.01, t(11)=2.28, p<0.05
(2) Binding of E. coli to cellulose powder
The results of F test and T test between c-1 and c-2; F(8,9)=33.88, p<0.01, t(8)=3.24, p<0.05
The results of F test and T test between d-1 and d-2; F(10,10)=18.64, p<0.01, t(11)=3.44, p<0.01</div>
Fig3 shows the results. After incubating E. coli with about 35~38 micrometer cellulose powder, we DAPI stained E. coli and observed it under fluorescence microscopy. As shown, CBD-surface expressing E. coli was observed binding to cellulose, showing the functional expression of INPNC-His-CBD in our system. When control plasmid was used, the numbers of cellulose-bound E. coli cells were significantly reduced (Fig4).
From these results, we concluded that this part can be used to physically link E. coli and cellulose.
Judging
We fulfilled criteria listed below with this part.
- Validated Part/ Validated Contribution
- [http://2016.igem.org/Team:Kyoto/HP/Gold Integrated Human Practice]
- Improve a previous part or project
- [http://2016.igem.org/Team:Kyoto/Proof Proof of concept]