Difference between revisions of "Part:BBa K1907002"
| Line 7: | Line 7: | ||
<p><b>Introduction</b></p> | <p><b>Introduction</b></p> | ||
This part can be used to introduce microcystinase (MlrA) from <i>Sphingomonas </i> sp. ACM-3962 to <i>Escherichia coli</i> BL21(DE3). MlrA is an enzyme degrades some cyanobacterial toxins, most notably microcystin-LR (MC-LR). To facilitate expression detection and purification of the enzyme, a 6X Histidine tag is fused on the C-terminus of MlrA, with a short linker in between. The full protein-coding sequence has been codon-optimized for expression in <i>E. coli</i> by GeneArt. | This part can be used to introduce microcystinase (MlrA) from <i>Sphingomonas </i> sp. ACM-3962 to <i>Escherichia coli</i> BL21(DE3). MlrA is an enzyme degrades some cyanobacterial toxins, most notably microcystin-LR (MC-LR). To facilitate expression detection and purification of the enzyme, a 6X Histidine tag is fused on the C-terminus of MlrA, with a short linker in between. The full protein-coding sequence has been codon-optimized for expression in <i>E. coli</i> by GeneArt. | ||
| + | <br> | ||
| + | This BioBrick is an improvement on the one created by the Peking 2014 team (BBa_K1378001). The MlrA sequence is completely identical to the MlrA sequence from <i>Sphingomonas</i> sp. ACM-3962 entry on GenBank (accession number AF411068.2), whereas the sequence of the older BioBrick differed by one amino acid. In the new BioBrick there is a 6X His-tag connected to the C-terminus by a linker. This tag will allow for the enzyme to purified by affinity chromatography and also detected by anti-His tag antibodies in western blotting. | ||
<br> | <br> | ||
Revision as of 00:32, 20 October 2016
Microcystinase (MlrA) for E. coli
Introduction
This part can be used to introduce microcystinase (MlrA) from Sphingomonas sp. ACM-3962 to Escherichia coli BL21(DE3). MlrA is an enzyme degrades some cyanobacterial toxins, most notably microcystin-LR (MC-LR). To facilitate expression detection and purification of the enzyme, a 6X Histidine tag is fused on the C-terminus of MlrA, with a short linker in between. The full protein-coding sequence has been codon-optimized for expression in E. coli by GeneArt.
This BioBrick is an improvement on the one created by the Peking 2014 team (BBa_K1378001). The MlrA sequence is completely identical to the MlrA sequence from Sphingomonas sp. ACM-3962 entry on GenBank (accession number AF411068.2), whereas the sequence of the older BioBrick differed by one amino acid. In the new BioBrick there is a 6X His-tag connected to the C-terminus by a linker. This tag will allow for the enzyme to purified by affinity chromatography and also detected by anti-His tag antibodies in western blotting.
Background
See BioBrick entry for MlrA in yeast (BBa_K1907001)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 105
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 244
Illegal AgeI site found at 373 - 1000COMPATIBLE WITH RFC[1000]
Expression
To express the enzyme, a 5 mL overnight-grown LB-kanamycin culture of E. coli BL21(DE3) was expanded to 50 mL. After the bacteria reached the mid-log phase (OD600 between 0.6-0.8), protein production was induced with 1 mM IPTG. During induction, bacteria were grown at +22 °C for 20 hours. Figure 1 presents results from analyzing different cellular fractions over the course of expression and lysis. After cell lysis, the enzyme is observed to localize in both the soluble and insoluble fractions.
Expression of the protein was also done in Magic Media auto-induction medium, in a total volume of 0.5 L in a 2 L flask covered with an air-permeable membrane. This expression was conducted in +30 °C.
FPLC purification
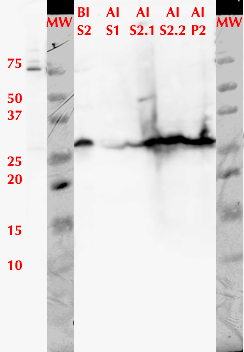
We used FPLC to purify the enzyme we had grown on the Magic Media auto-induction medium (0.5 L culture volume). Half of the supernatant obtained after cell lysis was used for purification. Figure 2 presents the chromatogram from the FPLC and selected fractions were analysed by SDS-PAGE and western blotting (Figure 3).
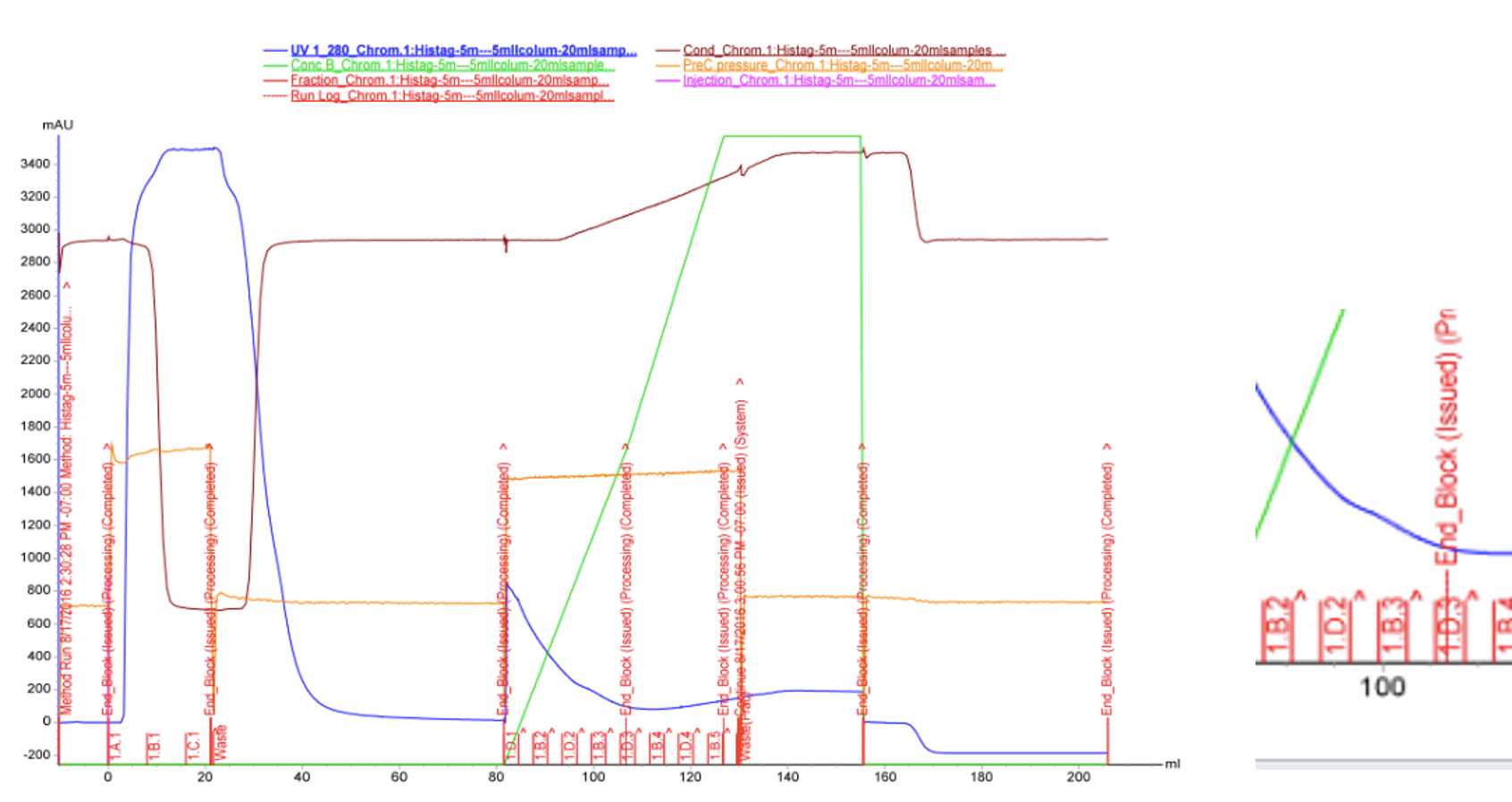
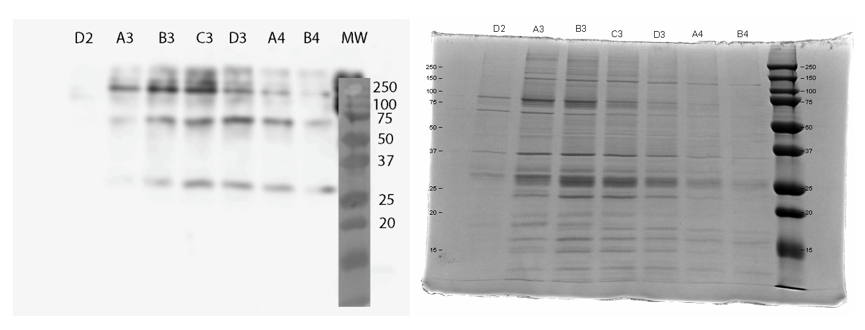
Based on these results, it seems that affinity chromatography purification of MlrA is possible, although optimization of the purification conditions would be necessary to obtain better purity and higher yields of purified protein. We didn’t have time to try these improvements ourselves, but possibilities include a very slow initial elution buffer gradient to wash out unspecific binding. In addition, optimizing the pH of the sample could improve binding; a higher pH might allow for better binding to the nickel in the column, as the number of histidines with a complementary charge would increase. Diluting the sample in phosphate buffer and desalting the sample could improve its binding to the column. Additionally, binding could be facilitated by extending the 6X His tag to a 10X His tag.
Activity measurements
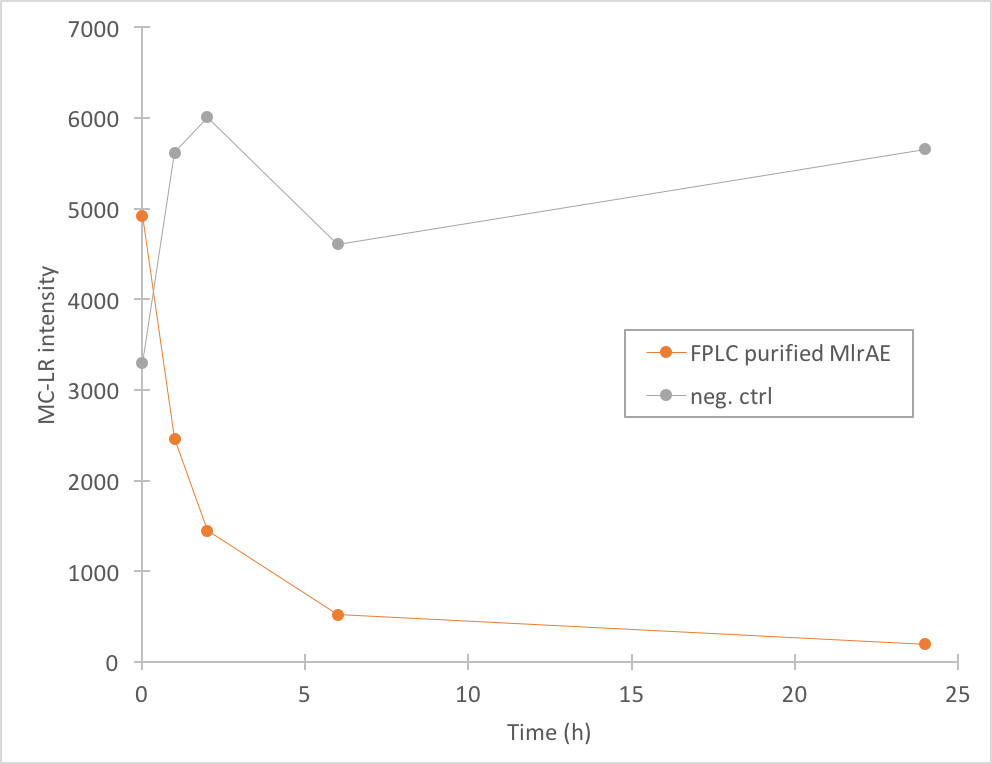
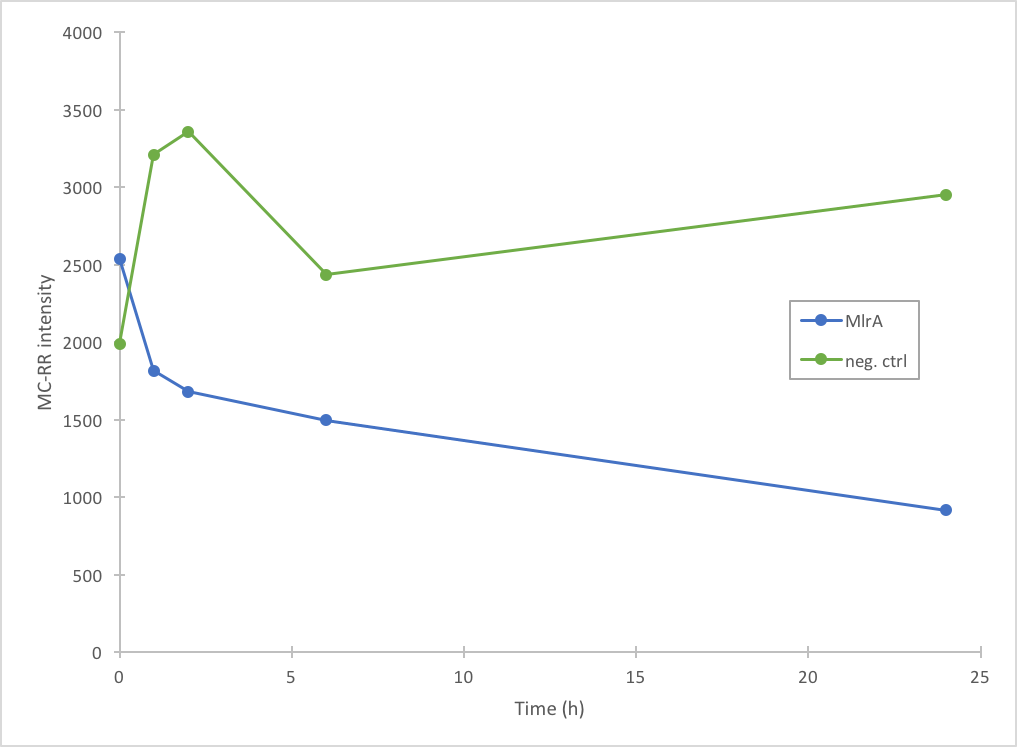
The activity of MlrA was measured by incubating the enzyme with microcystin extract. The final concentration of microcystin was estimated to be 1 μg/L. Samples were taken at different time points and they were analysed with liquid chromatography/mass spectrometry (LC-MS). This gives data of the relative abundance of microcystin-LR and -RR in each sample. The results were then plotted against time, together with a negative control which consisted of microcystin extract and PBS.
The results we acquired from this construct show that the amount of microcystin-LR decreases, which means that the enzyme is active (Figure 4). We used 970 ul of FPLC purified elution fraction in the MlrA-MC reaction, but as we don’t have exact information on the quantity of the enzyme, we were not able to quantify the activity activity any further.
In addition to improving the previous MlrA BioBrick, we also managed to prove that our construct degrades microcystin-RR (Figure 5). Based on our results, it seems that MlrA has higher degradation activity against MC-LR than MC-RR.