Difference between revisions of "Part:BBa K2110105"
(→Results) |
(→Results) |
||
| Line 11: | Line 11: | ||
===Sequence and Features=== | ===Sequence and Features=== | ||
<partinfo>BBa_K2110105 SequenceAndFeatures</partinfo> | <partinfo>BBa_K2110105 SequenceAndFeatures</partinfo> | ||
| − | = | + | <div class="row"> |
| − | <h2>Rational Design</ | + | <h2><b>Protein Engineering</b></h2> |
| − | <h3>Serine-based Catalytic Triad Mechanism & 3D Model Simulation</h3> | + | <h3><b>Outline</b></h3> |
| − | Since PETase was found to contain a <b>GWSMG</b> motif in accordance with the <b>GXGXG</b> motif | + | <p style="font-size:18px">The real goal of the protein engineering part in our project was to create a more active form of PETase protein. We designed and expressed 22 mutations in cell-free protein synthesis system, a brand new way to do the high-throughout assay. And from the 22 mutations, we successfully screened out a mutation I208V with hydrolysis activity improved by two-fold compared to the wild-type PETase.</p> |
| − | <br/>https://static.igem.org/mediawiki/2016/5/57/Fig.1.PNG | + | |
| − | < | + | <h3><b>Rational Design</b></h3> |
| − | < | + | <h3>Serine-based Catalytic Triad Mechanism & 3D Model Simulation</h3> |
| − | <h3>Mutation Design</h3> | + | <p style="font-size:18px">Since PETase was found to contain a <b>GWSMG</b> motif in accordance with the <b>GXGXG</b> motif, which is characteristic and highly conserved in α/β hydrolase fold family, we simulated a best fit model for PETase by SWISSMODEL with a template as Thc_Cut2. As expected, the homology model of PETase displays a canonical α/β hydrolase fold with a Ser<sup>160</sup>-His<sup>237</sup>-Asp<sup>206</sup> catalytic triad and a preformed oxyanion hole (Fig.1), suggesting a classic serine hydrolase mechanism. |
| − | Based on the simulated 3D structure, we found the 208th (with signal peptide excluded) amino acid residue isoleucine situates right over the catalytic triad(Fig.2). It is apperant that the interaction of catalytic triad with substrate would be inhibited due to the space oppcupied by 208th isoleucine. So we decided to substitue 208th isoleucine with another smaller amino acid. | + | </p><br/><br/> |
| − | <br/> | + | <div class="col-md-2"></div> |
| − | + | <div class="col-md-8"> | |
| − | <br/> | + | <div align="center"> |
| − | However, as the isoleucine is the most hydrophobic amino acid, it takes risk to substitue it with another one, as it may lead to the hydrophobic amino acid decreasing. On balance, we chose valine as substitution, for its hydrophobicity is quite close to isoleucine, and the volume of valine is much smaller than that of isoleucine. | + | <figure><a href="https://static.igem.org/mediawiki/2016/5/57/Fig.1.PNG" data-lightbox="no" data-title=">Fig.1 Simulated 3D structure for PETase"><img src="https://static.igem.org/mediawiki/2016/5/57/Fig.1.PNG" ></a><figcation>Fig.1 Simulated 3D structure for PETase</figcation></figure></div> </div> |
| − | <br/><br/> | + | <div class="col-md-2"></div> |
| − | The 3D structure of mutant I208V is as Fig.3. Comparing to wild-type PETase | + | |
| − | https://static.igem.org/mediawiki/2016/7/78/V208.png | + | |
| − | < | + | <div class="col-md-12"> |
| + | <h3>Mutation Design</h3> | ||
| + | <p style="font-size:18px">Based on the simulated 3D structure, we found the 208th (with signal peptide excluded) amino acid residue isoleucine situates right over the catalytic triad(Fig.2). It is apperant that the interaction of catalytic triad with substrate would be inhibited due to the space oppcupied by 208th isoleucine. So we decided to substitue 208th isoleucine with another smaller amino acid. | ||
| + | <br/><br/> | ||
| + | However, as the isoleucine is the most hydrophobic amino acid, it takes risk to substitue it with another one, as it may lead to the hydrophobic amino acid decreasing. On balance, we chose valine as substitution, for its hydrophobicity is quite close to isoleucine, and the volume of valine is much smaller than that of isoleucine. | ||
| + | <br/><br/> | ||
| + | The 3D structure of mutant I208V is as Fig.3. Comparing to wild-type PETase, the mutant I208V exposes the catalytic triad more widely. And the assay shows the stratety worked well for improving PET hydrolysis activity. The results are as follows. | ||
| + | </p></div> | ||
| + | |||
| + | <div class="col-md-6"> | ||
| + | <div align="center"> | ||
| + | <figure><a href="https://static.igem.org/mediawiki/2016/9/99/I208.png" data-lightbox="no" data-title="Fig.2 PETase Structure with 208<sup>th</sup>Isoleucine"><img src="https://static.igem.org/mediawiki/2016/9/99/I208.png" ></a><figcation>Fig.2 PETase Structure with 208<sup>th</sup>Isoleucine</figcation></figure></div> </div> | ||
| + | |||
| + | <div class="col-md-6"> | ||
| + | <div align="center"> | ||
| + | <figure><a href="https://static.igem.org/mediawiki/2016/7/78/V208.png" data-lightbox="no" data-title="Fig.3 PETase Structure with 208<sup>th</sup> Valine"><img src="https://static.igem.org/mediawiki/2016/7/78/V208.png" ></a><figcation>Fig.3 PETase Structure with 208<sup>th</sup> Valine</figcation></figure></div> </div> | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <div class="row"> | ||
| + | <div class="col-md-12"> | ||
| + | <h3>Plasmid construction and expression in CFPS system</h3> | ||
| + | <p style="font-size:18px">Ligaion of digested pRset_CFP-1, digested CFP gene and digested PETase(I208V) gene in accordance with the 1:5:5 molecular ratio. The newly constructed plasmid is called pRset_CFP-1_PETase-I208V.Then, the plasmids pRset_CFP-1_PETase-I208V was put into the CFPS(Cell-Free Protein Synthesis) to synthesis the enzymes we expected. </p> | ||
| + | <div class="col-md-2"></div> | ||
| + | <div class="col-md-8"> | ||
| + | <div align="center"> | ||
| + | <figure><a href="https://static.igem.org/mediawiki/2016/e/e6/T--Tianjin--part-I208Vpc.png" data-lightbox="no" data-title=">Fig.4 Plasmid construction and expression in CFPS ststem"><img src="https://static.igem.org/mediawiki/2016/e/e6/T--Tianjin--part-I208Vpc.png" ></a><figcation>Fig.4 Plasmid construction and expression in CFPS ststem</figcation></figure></div> </div> | ||
| + | <div class="col-md-2"></div> | ||
| + | </div></div> | ||
| − | + | ||
| − | < | + | |
| − | <h3> | + | <div class="row"> |
| − | + | ||
| + | <h3>Degradation data</h3> | ||
| + | <p style="font-size:18px">After the enzymes were expressed in the system successfully, we used the mutants we got to degrade PET and detected the degradation product, MHET, which has no other characteristic adsorption peak except at 260nm. | ||
| − | + | We detect the absorption peak of culture medium after culturing with PET film added for 5d. The absorption degree-wavelength curve is as follows.</p> | |
| + | |||
| + | |||
| + | <div class="col-md-2"></div> | ||
| + | <div class="col-md-8"> | ||
| + | <div align="center"> | ||
| + | <figure><a href="https://static.igem.org/mediawiki/parts/thumb/5/5a/T--Tianjin--i208vnew.png/800px-T--Tianjin--i208vnew.png" data-lightbox="no" data-title=">Fig.5 Spectra scanning for the degradation product"><img src="https://static.igem.org/mediawiki/parts/thumb/5/5a/T--Tianjin--i208vnew.png/800px-T--Tianjin--i208vnew.png" style="height:500px"></a><figcation>Fig.5 Spectra scanning for the degradation product</figcation></figure></div> </div> | ||
| + | <div class="col-md-2"></div></div> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | ||
| − | + | ||
| − | < | + | <div class="row"> |
| − | https://static.igem.org/mediawiki/ | + | <p style="font-size:18px">We used Cyan fluorescence protein (CFP) as a real-time approach to monitor protein expression quantity by detecting the fluorescence signal in the plate-reader. The emission wavelength of the CFP is 479nm and the absorption wavelength is 435nm. This is the real-time detection of emission wavelength at 479nm. We detected the fluorescence absorption for 12 hours before adding PET film substrate.</p> |
| + | <div class="col-md-2"></div> | ||
| + | <div class="col-md-8"> | ||
| + | <div align="center"> | ||
| + | <figure><a href="https://static.igem.org/mediawiki/2016/1/1d/T--Tianjin--part-I208Vex.png" data-lightbox="no" data-title="Fig.6 Screened plasmids expressions in the CFPS system"><img src="https://static.igem.org/mediawiki/2016/1/1d/T--Tianjin--part-I208Vex.png" ></a><figcation>Fig.6 Screened plasmids expressions in the CFPS system</figcation></figure></div> </div> | ||
| + | <div class="col-md-2"></div> | ||
| + | </div> | ||
| − | < | + | <div class="row"> |
| + | <p style="font-size:18px">To exclude the influence of different expression quantity, we divided the quantity of production of PET degradation of each mutant by its expression quantity to gain the relative activity, since the final concentration of product is positively relevant to absorption at 260nm and protein expression quantity is positively relevant to the absorption of CFP at emission wavelength.</p></div> | ||
| − | + | ||
| + | <div class="row"> | ||
| + | <div class="col-md-2"></div> | ||
| + | <div class="col-md-8"> | ||
| + | <div align="center"> | ||
| + | <figure> | ||
| + | <a href="https://static.igem.org/mediawiki/2016/2/2f/T--Tianjin--part-I208Vbar.png" data-lightbox="no" data-title="Fig.7 Relative enzyme activity of Plasmid pRset_CFP-1_PETase-I208V"><img src="https://static.igem.org/mediawiki/2016/2/2f/T--Tianjin--part-I208Vbar.png" ></a> | ||
| + | <figcation>Fig.7 Relative enzyme activity of Plasmid pRset_CFP-1_PETase-I208V<br/></figcation></figure></div></div> | ||
| + | <div class="col-md-2"></div> | ||
| − | + | <div class="col-md-12"> | |
| − | < | + | <p style="font-size:18px">So our result shows the rational design actually worked well and the mutant I208V shows a relatively improved hyfrolysis activity comparing to wild-type. Though as we can see in Fig.3, the substitution of isoleucine with valine is actually a very small change with a methyl on the 208<sup>th</sup> amino residue. However it leads to a two-fold improvement of enzyme activity, revealing that as we expected the 208<sup>th</sup> is a crucial site and the space adjacent to active center matters a lot. </p> |
| − | + | </div> | |
| − | + | </div> | |
| − | + | ||
| − | < | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
Revision as of 22:29, 19 October 2016
PETase site-directed mutant I208V
This is one of the site-directed mutants of the PETase we designed, we change the 208th amino acid isoleucine(I) with valine(V). The module is the Saccharomyces cerevisiae codon optimization version of PETase gene. We used site-directed mutagenesis PCR to obtain this mutant.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 79
Illegal BglII site found at 289 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Protein Engineering
Outline
The real goal of the protein engineering part in our project was to create a more active form of PETase protein. We designed and expressed 22 mutations in cell-free protein synthesis system, a brand new way to do the high-throughout assay. And from the 22 mutations, we successfully screened out a mutation I208V with hydrolysis activity improved by two-fold compared to the wild-type PETase.
Rational Design
Serine-based Catalytic Triad Mechanism & 3D Model Simulation
Since PETase was found to contain a GWSMG motif in accordance with the GXGXG motif, which is characteristic and highly conserved in α/β hydrolase fold family, we simulated a best fit model for PETase by SWISSMODEL with a template as Thc_Cut2. As expected, the homology model of PETase displays a canonical α/β hydrolase fold with a Ser160-His237-Asp206 catalytic triad and a preformed oxyanion hole (Fig.1), suggesting a classic serine hydrolase mechanism.
 " data-lightbox="no" data-title=">Fig.1 Simulated 3D structure for PETase"><img src="
" data-lightbox="no" data-title=">Fig.1 Simulated 3D structure for PETase"><img src=" " ></a><figcation>Fig.1 Simulated 3D structure for PETase</figcation></figure>
" ></a><figcation>Fig.1 Simulated 3D structure for PETase</figcation></figure>
Mutation Design
Based on the simulated 3D structure, we found the 208th (with signal peptide excluded) amino acid residue isoleucine situates right over the catalytic triad(Fig.2). It is apperant that the interaction of catalytic triad with substrate would be inhibited due to the space oppcupied by 208th isoleucine. So we decided to substitue 208th isoleucine with another smaller amino acid.
However, as the isoleucine is the most hydrophobic amino acid, it takes risk to substitue it with another one, as it may lead to the hydrophobic amino acid decreasing. On balance, we chose valine as substitution, for its hydrophobicity is quite close to isoleucine, and the volume of valine is much smaller than that of isoleucine.
The 3D structure of mutant I208V is as Fig.3. Comparing to wild-type PETase, the mutant I208V exposes the catalytic triad more widely. And the assay shows the stratety worked well for improving PET hydrolysis activity. The results are as follows.
 " data-lightbox="no" data-title="Fig.2 PETase Structure with 208thIsoleucine"><img src="
" data-lightbox="no" data-title="Fig.2 PETase Structure with 208thIsoleucine"><img src=" " ></a><figcation>Fig.2 PETase Structure with 208thIsoleucine</figcation></figure>
" ></a><figcation>Fig.2 PETase Structure with 208thIsoleucine</figcation></figure>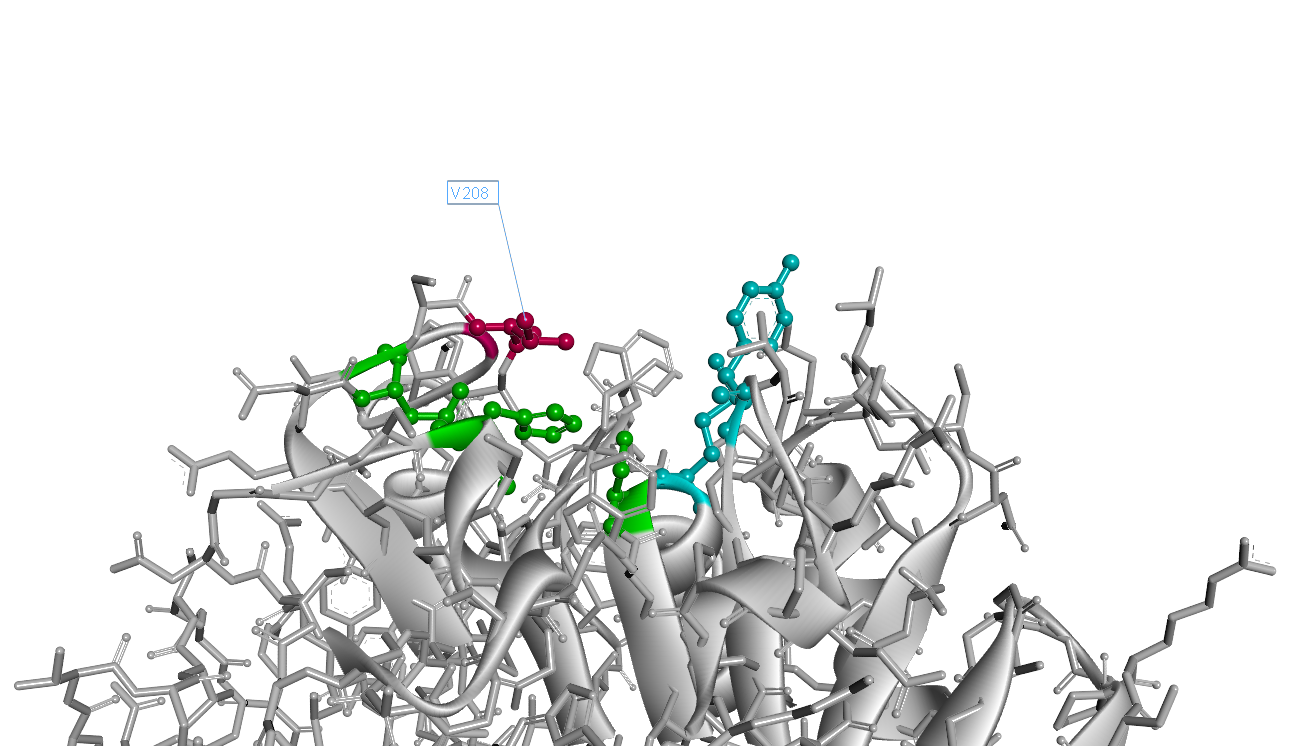 " data-lightbox="no" data-title="Fig.3 PETase Structure with 208th Valine"><img src="
" data-lightbox="no" data-title="Fig.3 PETase Structure with 208th Valine"><img src=" " ></a><figcation>Fig.3 PETase Structure with 208th Valine</figcation></figure>
" ></a><figcation>Fig.3 PETase Structure with 208th Valine</figcation></figure>
Plasmid construction and expression in CFPS system
Ligaion of digested pRset_CFP-1, digested CFP gene and digested PETase(I208V) gene in accordance with the 1:5:5 molecular ratio. The newly constructed plasmid is called pRset_CFP-1_PETase-I208V.Then, the plasmids pRset_CFP-1_PETase-I208V was put into the CFPS(Cell-Free Protein Synthesis) to synthesis the enzymes we expected.
 " data-lightbox="no" data-title=">Fig.4 Plasmid construction and expression in CFPS ststem"><img src="
" data-lightbox="no" data-title=">Fig.4 Plasmid construction and expression in CFPS ststem"><img src=" " ></a><figcation>Fig.4 Plasmid construction and expression in CFPS ststem</figcation></figure>
" ></a><figcation>Fig.4 Plasmid construction and expression in CFPS ststem</figcation></figure>
Degradation data
After the enzymes were expressed in the system successfully, we used the mutants we got to degrade PET and detected the degradation product, MHET, which has no other characteristic adsorption peak except at 260nm. We detect the absorption peak of culture medium after culturing with PET film added for 5d. The absorption degree-wavelength curve is as follows.
 " data-lightbox="no" data-title=">Fig.5 Spectra scanning for the degradation product"><img src="
" data-lightbox="no" data-title=">Fig.5 Spectra scanning for the degradation product"><img src=" " style="height:500px"></a><figcation>Fig.5 Spectra scanning for the degradation product</figcation></figure>
" style="height:500px"></a><figcation>Fig.5 Spectra scanning for the degradation product</figcation></figure>
We used Cyan fluorescence protein (CFP) as a real-time approach to monitor protein expression quantity by detecting the fluorescence signal in the plate-reader. The emission wavelength of the CFP is 479nm and the absorption wavelength is 435nm. This is the real-time detection of emission wavelength at 479nm. We detected the fluorescence absorption for 12 hours before adding PET film substrate.
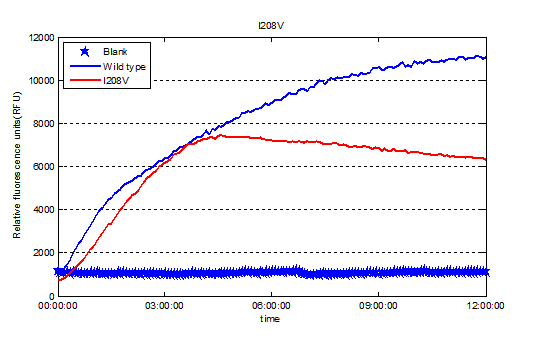 " data-lightbox="no" data-title="Fig.6 Screened plasmids expressions in the CFPS system"><img src="
" data-lightbox="no" data-title="Fig.6 Screened plasmids expressions in the CFPS system"><img src=" " ></a><figcation>Fig.6 Screened plasmids expressions in the CFPS system</figcation></figure>
" ></a><figcation>Fig.6 Screened plasmids expressions in the CFPS system</figcation></figure>
To exclude the influence of different expression quantity, we divided the quantity of production of PET degradation of each mutant by its expression quantity to gain the relative activity, since the final concentration of product is positively relevant to absorption at 260nm and protein expression quantity is positively relevant to the absorption of CFP at emission wavelength.
<figure>
<a href=" " data-lightbox="no" data-title="Fig.7 Relative enzyme activity of Plasmid pRset_CFP-1_PETase-I208V"><img src="
" data-lightbox="no" data-title="Fig.7 Relative enzyme activity of Plasmid pRset_CFP-1_PETase-I208V"><img src=" " ></a>
" ></a>
<figcation>Fig.7 Relative enzyme activity of Plasmid pRset_CFP-1_PETase-I208V</figcation></figure>
So our result shows the rational design actually worked well and the mutant I208V shows a relatively improved hyfrolysis activity comparing to wild-type. Though as we can see in Fig.3, the substitution of isoleucine with valine is actually a very small change with a methyl on the 208th amino residue. However it leads to a two-fold improvement of enzyme activity, revealing that as we expected the 208th is a crucial site and the space adjacent to active center matters a lot.
