Difference between revisions of "Part:BBa K1887003"
(→Design) |
(→Design) |
||
| (One intermediate revision by the same user not shown) | |||
| Line 27: | Line 27: | ||
[[file:cA1.jpg]] | [[file:cA1.jpg]] | ||
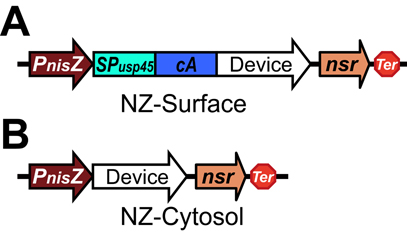
| − | Schematic of surface display system (A) and Cytosol express (B). Abbreviations: <i>PnisZ</i>, promoter; <i> | + | Schematic of surface display system (A) and Cytosol express (B). Abbreviations: <i>PnisZ</i>, promoter; <i>SP<sub>usp45</sub></i>, signal peptide of <i>usp45</i> from <i>L. lactis</i>; <i>cA</i>, anchor motif of <i>acmA</i> from <i>L. lactis</i>; <i>nsr</i>, nisin resistant gene; <i>Ter</i>, terminator. |
===Results=== | ===Results=== | ||
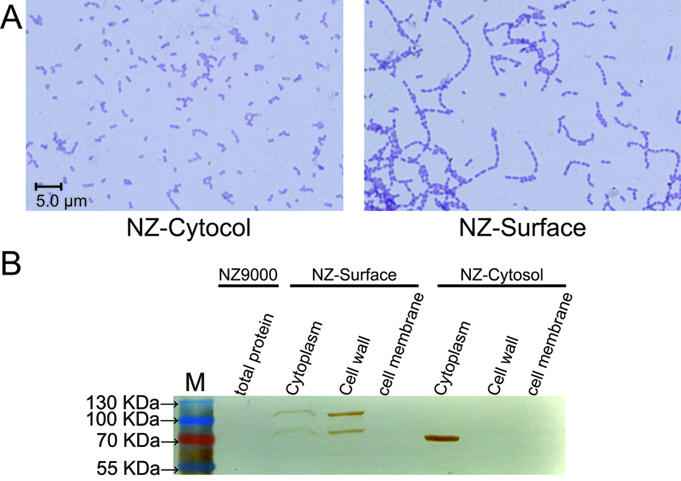
| − | To demonstrate whether cA could indeed anchor the protein of interest at the surface of | + | To demonstrate whether the SPusp45 signal peptide and the cA anchoring domain could indeed secret and anchor the protein of interest at the surface of <i>L. lactis</i>, the p<i>lacZ</i>-Cytosol and p<i>lacZ</i>-Surface plasmids were introduced into NZ9000 respectively, which were named NZ-Cytosol and NZ-Surface. As stated above, the cA domain come from the AcmA protein, which is an autolysin of <i>L. lactis</i> that cleaves the peptidoglycan to release the duplicated bacteria. Since the location sites of AcmA autolysin, substrate peptidoglycan, is now occupied by the β-galactosidase-cA fusion proteins secreted by SPusp45, the autolysin activity is hindered, thus cell separation will be interfered. Indeed, we found that under microscopic, the NZ-Surface cells were poorly separated compared to the NZ-Cytosol strain. Further more, using a polyclonal antibody against β-galactosidase, we found that the β-galactosidase protein is present at the cell wall and cytoplasm, which means the signal peptide, SPusp45, and anchor motif, cA domain, worked as we designed. In contrast, all the β-galactosidase proteins were present in the cytoplasm in the NZ-cytosol strain. |
[[File:cA2.jpg]] | [[File:cA2.jpg]] | ||
| − | The functional verification of NZ-Cytosol and NZ-Surface. A). Microscopic picture of the NZ-Cytosol and NZ-surface | + | The functional verification of the NZ-Cytosol and NZ-Surface strains. A). Microscopic picture of the NZ-Cytosol and NZ-surface strains. B). Western blot of different fractions from NZ9000, NZ-Cytosol and NZ-Surface. The protein fused with cA motif was located on the bacterial cell wall and the cytosol expressed protein was located in the bacterial cytoplasm. This mean the protein fused with cA motif was secreted by SPusp45 and was anchored in the cell wall by cA motif. The <i>lacZ</i> was used as an example. The strain used in western blot are presented upon the lanes |
===References=== | ===References=== | ||
Latest revision as of 17:39, 19 October 2016
cA anchoring domain
A cell wall binding domain used in surface display for L. lactis
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Molecular verification
We used PCR and restriction enzyme digestion to verify the cA part on pSB1C3.
Background
To make protein of interest to be cell surface displayed, anchoring motif should be considered at the first. The most exploited anchoring regions are those with the LPXTG motifs that bind the proteins in a covalent way to the cell wall. However, to achieve versatile application, it is better to use anchoring domains that interact with the cell wall in a non-covalent way. The major autolysin of L. lactis, the cell wall hydrolase AcmA contains 3 tandem arranged LysM motifs and separated by stretches of 21 to 31 amino acids, this region is collectively termed as the cA anchoring domain. The cA domain can be fused to the N- and C- terminus of functional proteins, and can bind proteins to the cell walls of a broad range of gram-positive bacteria.
Design
The protein of interest is fused to the cA domain with the USP45 signal peptide, driven by the PnisZ promoter and followed by the nisin resistant gene nsr.
Schematic of surface display system (A) and Cytosol express (B). Abbreviations: PnisZ, promoter; SPusp45, signal peptide of usp45 from L. lactis; cA, anchor motif of acmA from L. lactis; nsr, nisin resistant gene; Ter, terminator.
Results
To demonstrate whether the SPusp45 signal peptide and the cA anchoring domain could indeed secret and anchor the protein of interest at the surface of L. lactis, the placZ-Cytosol and placZ-Surface plasmids were introduced into NZ9000 respectively, which were named NZ-Cytosol and NZ-Surface. As stated above, the cA domain come from the AcmA protein, which is an autolysin of L. lactis that cleaves the peptidoglycan to release the duplicated bacteria. Since the location sites of AcmA autolysin, substrate peptidoglycan, is now occupied by the β-galactosidase-cA fusion proteins secreted by SPusp45, the autolysin activity is hindered, thus cell separation will be interfered. Indeed, we found that under microscopic, the NZ-Surface cells were poorly separated compared to the NZ-Cytosol strain. Further more, using a polyclonal antibody against β-galactosidase, we found that the β-galactosidase protein is present at the cell wall and cytoplasm, which means the signal peptide, SPusp45, and anchor motif, cA domain, worked as we designed. In contrast, all the β-galactosidase proteins were present in the cytoplasm in the NZ-cytosol strain.
The functional verification of the NZ-Cytosol and NZ-Surface strains. A). Microscopic picture of the NZ-Cytosol and NZ-surface strains. B). Western blot of different fractions from NZ9000, NZ-Cytosol and NZ-Surface. The protein fused with cA motif was located on the bacterial cell wall and the cytosol expressed protein was located in the bacterial cytoplasm. This mean the protein fused with cA motif was secreted by SPusp45 and was anchored in the cell wall by cA motif. The lacZ was used as an example. The strain used in western blot are presented upon the lanes
References
[1] Buist, G., Steen, A., Kok, J., and Kuipers, O.P. (2008). LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68, 838-847.
[2] Raha, A.R., Varma, N.R., Yusoff, K., Ross, E., and Foo, H.L. (2005). Cell surface display system for Lactococcus lactis: a novel development for oral vaccine. Appl Microbiol Biotechnol 68, 75-81.