Difference between revisions of "Part:BBa K1654007"
(→Design) |
(→Usage and Biology) |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K1654007 short</partinfo> | <partinfo>BBa_K1654007 short</partinfo> | ||
| − | USP45 secretion tag used in Lactococcus lactis to secrete the protein outside the cell. | + | USP45 secretion tag used in ''Lactococcus lactis'' to secrete the protein outside the cell. |
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 12: | Line 12: | ||
<partinfo>BBa_K1654007 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1654007 SequenceAndFeatures</partinfo> | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | The [http://2016.igem.org/Team:TMMU_China/Project_Results/Parts TMMU-China 2016 iGEM team] verified this part, see below. | ||
| + | |||
===Background=== | ===Background=== | ||
| − | To get the protein of interest anchored to the surface of L. lactis, it should be secreted out of the cell first. The solution is a signal peptide from the Usp45 protein which is secreted out of L. lactis very efficiently. Fusion of the signal peptide of Usp45 to a couple of proteins resulted in an efficient way of secreting the coded proteins into the media. The fusion protein will meet a chaperone protein, SecB, which bring the fusion protein to a protein secretion apparatus. This apparatus cleaves the signal peptide and frees the proteins from the cell. | + | To get the protein of interest anchored to the surface of ''L. lactis'', it should be secreted out of the cell first. The solution is a signal peptide from the Usp45 protein which is secreted out of L. lactis very efficiently. Fusion of the signal peptide of Usp45 to a couple of proteins resulted in an efficient way of secreting the coded proteins into the media. The fusion protein will meet a chaperone protein, SecB, which bring the fusion protein to a protein secretion apparatus. This apparatus cleaves the signal peptide and frees the proteins from the cell. |
===Design=== | ===Design=== | ||
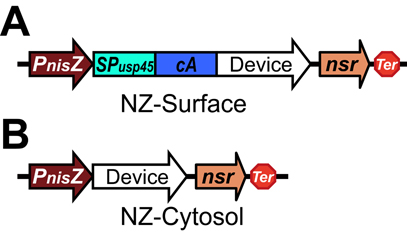
The protein of interest is fused to the cA domain with the USP45 signal peptide, driven by the PnisZ promoter and followed by the nisin resistant gene nsr. To demonstrate the utility of the protein surface display, here we took the β-galactosidase protein as a proof of concept. | The protein of interest is fused to the cA domain with the USP45 signal peptide, driven by the PnisZ promoter and followed by the nisin resistant gene nsr. To demonstrate the utility of the protein surface display, here we took the β-galactosidase protein as a proof of concept. | ||
[[file:cA1.jpg]] | [[file:cA1.jpg]] | ||
| + | |||
| + | Schematic of surface display system (A) and Cytosol express (B). Abbreviations: PnisZ, promoter; SPusp45, signal peptide of usp45 from L. lactis; cA, anchor motif of acmA from L. lactis; nsr, nisin resistant gene; Ter, terminator. | ||
===Results=== | ===Results=== | ||
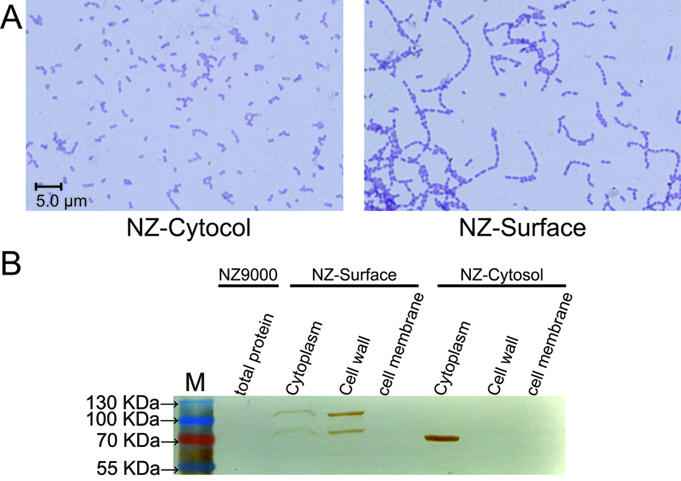
| − | To demonstrate whether cA could indeed anchor the protein of interest at the surface of L. lactis, the pLacZ-Cytosol and pLacZ-Surface plasmids were introduced into NZ9000 respectively. As stated above, the cA domain is derived from the AcmA protein, which is an autolysin of L. latis that cleaves the peptidoglycan to release the duplicated bacteria. Since the substrate peptidoglycan is now occupied by the cA-β-galactosidase fusion proteins, the AcmA autolysin activity is hindered, thus cell separation will be interfered. Indeed, we found that under microscopic, the NZ-Surface cells were poorly separated compared to the NZ-Cytosol strain. Further more, using a polyclonal antibody against β-galactosidase, we found that about 70% of the β-galactosidase protein is present at the cell wall, while the other 30% protein is present in the cytoplasm, perhaps due to the inefficient secretion process. In contrast, all theβ-galactosidase proteins were present in the cytoplasm in the NZ-cytosol strain. | + | To demonstrate whether cA could indeed anchor the protein of interest at the surface of ''L. lactis'', the pLacZ-Cytosol and pLacZ-Surface plasmids were introduced into NZ9000 respectively. As stated above, the cA domain is derived from the AcmA protein, which is an autolysin of L. latis that cleaves the peptidoglycan to release the duplicated bacteria. Since the substrate peptidoglycan is now occupied by the cA-β-galactosidase fusion proteins, the AcmA autolysin activity is hindered, thus cell separation will be interfered. Indeed, we found that under microscopic, the NZ-Surface cells were poorly separated compared to the NZ-Cytosol strain. Further more, using a polyclonal antibody against β-galactosidase, we found that about 70% of the β-galactosidase protein is present at the cell wall, while the other 30% protein is present in the cytoplasm, perhaps due to the inefficient secretion process. In contrast, all theβ-galactosidase proteins were present in the cytoplasm in the NZ-cytosol strain. |
[[File:cA2.jpg]] | [[File:cA2.jpg]] | ||
| − | The | + | The functional verification of NZ-Cytosol and NZ-Surface. A). Microscopic picture of the NZ-Cytosol and NZ-surface. The anchor motif cA is come from the autolysins AcmA of L. lactis. Protein fused with cA motif will compete with the autolysins AcmA and inhibited the separated of the divide bacteria cells, which makes NZ-surface strain grow in longer chains than NZ-Cytosol strain. B). Western blot of different fractions from NZ9000, NZ-Cytosol and NZ-Surface. The protein fused with cA motif was located on the bacterial cell wall and the cytosol expressed protein was located in the bacterial cytoplasm. This mean the protein fused with cA motif was secreted by SPusp45 and was anchored in the cell wall by cA motif. The LacZ was used as an example. The strain used in western blot are presented upon the lanes |
| − | + | ||
| + | ===Reference=== | ||
| + | Le Loir, Y., Nouaille, S., Commissaire, J., Bretigny, L., Gruss, A., and Langella, P. (2001). Signal peptide and propeptide optimization for heterologous protein secretion in Lactococcus lactis. Appl Environ Microbiol 67, 4119-4127. | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 03:37, 17 October 2016
USP45 secretion tag for Lactococcus lactis
USP45 secretion tag used in Lactococcus lactis to secrete the protein outside the cell.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
The [http://2016.igem.org/Team:TMMU_China/Project_Results/Parts TMMU-China 2016 iGEM team] verified this part, see below.
Background
To get the protein of interest anchored to the surface of L. lactis, it should be secreted out of the cell first. The solution is a signal peptide from the Usp45 protein which is secreted out of L. lactis very efficiently. Fusion of the signal peptide of Usp45 to a couple of proteins resulted in an efficient way of secreting the coded proteins into the media. The fusion protein will meet a chaperone protein, SecB, which bring the fusion protein to a protein secretion apparatus. This apparatus cleaves the signal peptide and frees the proteins from the cell.
Design
The protein of interest is fused to the cA domain with the USP45 signal peptide, driven by the PnisZ promoter and followed by the nisin resistant gene nsr. To demonstrate the utility of the protein surface display, here we took the β-galactosidase protein as a proof of concept.
Schematic of surface display system (A) and Cytosol express (B). Abbreviations: PnisZ, promoter; SPusp45, signal peptide of usp45 from L. lactis; cA, anchor motif of acmA from L. lactis; nsr, nisin resistant gene; Ter, terminator.
Results
To demonstrate whether cA could indeed anchor the protein of interest at the surface of L. lactis, the pLacZ-Cytosol and pLacZ-Surface plasmids were introduced into NZ9000 respectively. As stated above, the cA domain is derived from the AcmA protein, which is an autolysin of L. latis that cleaves the peptidoglycan to release the duplicated bacteria. Since the substrate peptidoglycan is now occupied by the cA-β-galactosidase fusion proteins, the AcmA autolysin activity is hindered, thus cell separation will be interfered. Indeed, we found that under microscopic, the NZ-Surface cells were poorly separated compared to the NZ-Cytosol strain. Further more, using a polyclonal antibody against β-galactosidase, we found that about 70% of the β-galactosidase protein is present at the cell wall, while the other 30% protein is present in the cytoplasm, perhaps due to the inefficient secretion process. In contrast, all theβ-galactosidase proteins were present in the cytoplasm in the NZ-cytosol strain.
The functional verification of NZ-Cytosol and NZ-Surface. A). Microscopic picture of the NZ-Cytosol and NZ-surface. The anchor motif cA is come from the autolysins AcmA of L. lactis. Protein fused with cA motif will compete with the autolysins AcmA and inhibited the separated of the divide bacteria cells, which makes NZ-surface strain grow in longer chains than NZ-Cytosol strain. B). Western blot of different fractions from NZ9000, NZ-Cytosol and NZ-Surface. The protein fused with cA motif was located on the bacterial cell wall and the cytosol expressed protein was located in the bacterial cytoplasm. This mean the protein fused with cA motif was secreted by SPusp45 and was anchored in the cell wall by cA motif. The LacZ was used as an example. The strain used in western blot are presented upon the lanes
Reference
Le Loir, Y., Nouaille, S., Commissaire, J., Bretigny, L., Gruss, A., and Langella, P. (2001). Signal peptide and propeptide optimization for heterologous protein secretion in Lactococcus lactis. Appl Environ Microbiol 67, 4119-4127.