Difference between revisions of "Part:BBa K1980000"
| Line 5: | Line 5: | ||
Copper Storage protein 1 (Csp1) is a tetrameric copper storage protein found in the periplasm of Methylosinus trichosporium OB3b. We investigated whether this part could act as a copper chelator when expressed in E. coli. | Copper Storage protein 1 (Csp1) is a tetrameric copper storage protein found in the periplasm of Methylosinus trichosporium OB3b. We investigated whether this part could act as a copper chelator when expressed in E. coli. | ||
We modified the protein by adding a TAT signal peptide from the E. coli enzyme CueO in place of the native TAT sequence and added a C terminal his tag to aid purification. | We modified the protein by adding a TAT signal peptide from the E. coli enzyme CueO in place of the native TAT sequence and added a C terminal his tag to aid purification. | ||
| − | + | <!-- --> | |
| + | <span class='h3bb'>Sequence and Features:</span> | ||
| + | <partinfo>BBa_K1980000 SequenceAndFeatures</partinfo> | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| Line 16: | Line 18: | ||
<li>As <i> E. coli </i>naturally deals with copper toxicity by binding copper in the periplasm then exporting it, periplasmic proteins may reduce toxicity to the host. </li> | <li>As <i> E. coli </i>naturally deals with copper toxicity by binding copper in the periplasm then exporting it, periplasmic proteins may reduce toxicity to the host. </li> | ||
</ul> | </ul> | ||
| − | <p> | + | <p>Copper storage protein 1 is a protein discovered in a methane-oxidizing alphaproteobacterium called<i>Methylosinus trichosporium OB3b. </i> (OB3b here stands for “oddball” strain 3b). This bacterium has a high demand for copper for use in its particular methane monoxygenase enzyme. Vita et al. <sup>(1)</sup> discovered Csp1 in 2015, characterised the protein’s copper affinity and obtained crystal structures with and without copper. |
| − | + | </p> | |
| + | <p>(1) Nicolas Vita, Semeli Platsaki, Arnaud Basle, Stephen J. Allen, Neil G. Paterson, Andrew T. Crombie, J. Colin Murrell, Kevin J.Waldron & Christopher Dennison (2015) | ||
| + | “A four-helix bundle stores copper for methane oxidation”, Nature 525 issue 7567 pg. 140-143 doi:10.1038/nature14854</p> | ||
| + | <p> | ||
| + | Csp1 is a tetramer of four-helix bundles. Each monomer can bind up to 13 Cu(I) ions meaning that the tetramer binds a maximum of 52 copper ions. Vita et al crystallised Csp1 with and without copper bound. The copper is bound inside the pre-folded helical bundles by Cys residues in contrast to metallothioneins, which are unstructured until they fold around metal ion clusters. Vita et al. found an average copper affinity of approximately 1x10<sup>17</sup>M<sup>-1</sup>. | ||
</p> | </p> | ||
| − | |||
| − | |||
| − | |||
| + | <p> | ||
| + | Csp1 has a signal peptide targeting it to the twin arginine translocation pathway (TAT). This means that it is likely a periplasmic protein. However they also found cytoplasmic homologues in many species challenging their and our assumption that only copper storage occurs in the periplasm due to copper toxicity. | ||
| + | </p> | ||
| + | <p> | ||
| + | We codon optimised Csp1 to <i>E. coli</i> and replaced the original TAT sequence with a TAT sequence from the <i>E. coli</i> protein CueO, which is also involved in copper regulation. To get Csp1 from the shipping vector to the pBAD expression system for testing the TAT sequence had to be modified by the addition of a serine residue after the initiator methionine. Serine was chosen over other amino acid possibilities because other TAT sequences seemed to have serine in this location. | ||
| + | </p> | ||
| + | <img src="https://static.igem.org/mediawiki/2016/0/0e/Csp1_single_for_Chelators_page_Sam_Oxford_2016.png" width="50%"/> | ||
| + | ==Experience== | ||
| + | <p>We were unable however to detect copper chelation activity of Csp1 <i>in vivo</i> using a BCS absorbance assay. Modelling by our team suggested that this was because insufficient protein could be expressed to chelate the amount (units of μmols) needed to be detectable on the assay. </p> | ||
| + | <p>Using a version of this part with a C-terminal His tagged-sfGFP we purified this protein but were still unable to detect copper chelation with the assay in our purified extracts.</p> | ||
| + | <p> | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Revision as of 20:14, 16 October 2016
TAT Copper Storage Protein 1
Copper Storage protein 1 (Csp1) is a tetrameric copper storage protein found in the periplasm of Methylosinus trichosporium OB3b. We investigated whether this part could act as a copper chelator when expressed in E. coli. We modified the protein by adding a TAT signal peptide from the E. coli enzyme CueO in place of the native TAT sequence and added a C terminal his tag to aid purification. Sequence and Features:
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Our project aimed to detect and chelate dietary copper as a treatment for Wilson's Disease, a copper accumulation disorder. We decided that the ideal copper chelation protein would have these properties:
- Should be able to bind multiple copper ions per peptide to increase the efficient use of cell resources.
- They should be from the prokaryotic domain because eukaryotic proteins can have expression issues in Escherichia coli.
- As E. coli naturally deals with copper toxicity by binding copper in the periplasm then exporting it, periplasmic proteins may reduce toxicity to the host.
Copper storage protein 1 is a protein discovered in a methane-oxidizing alphaproteobacterium calledMethylosinus trichosporium OB3b. (OB3b here stands for “oddball” strain 3b). This bacterium has a high demand for copper for use in its particular methane monoxygenase enzyme. Vita et al. (1) discovered Csp1 in 2015, characterised the protein’s copper affinity and obtained crystal structures with and without copper.
(1) Nicolas Vita, Semeli Platsaki, Arnaud Basle, Stephen J. Allen, Neil G. Paterson, Andrew T. Crombie, J. Colin Murrell, Kevin J.Waldron & Christopher Dennison (2015) “A four-helix bundle stores copper for methane oxidation”, Nature 525 issue 7567 pg. 140-143 doi:10.1038/nature14854
Csp1 is a tetramer of four-helix bundles. Each monomer can bind up to 13 Cu(I) ions meaning that the tetramer binds a maximum of 52 copper ions. Vita et al crystallised Csp1 with and without copper bound. The copper is bound inside the pre-folded helical bundles by Cys residues in contrast to metallothioneins, which are unstructured until they fold around metal ion clusters. Vita et al. found an average copper affinity of approximately 1x1017M-1.
Csp1 has a signal peptide targeting it to the twin arginine translocation pathway (TAT). This means that it is likely a periplasmic protein. However they also found cytoplasmic homologues in many species challenging their and our assumption that only copper storage occurs in the periplasm due to copper toxicity.
We codon optimised Csp1 to E. coli and replaced the original TAT sequence with a TAT sequence from the E. coli protein CueO, which is also involved in copper regulation. To get Csp1 from the shipping vector to the pBAD expression system for testing the TAT sequence had to be modified by the addition of a serine residue after the initiator methionine. Serine was chosen over other amino acid possibilities because other TAT sequences seemed to have serine in this location.
<img src="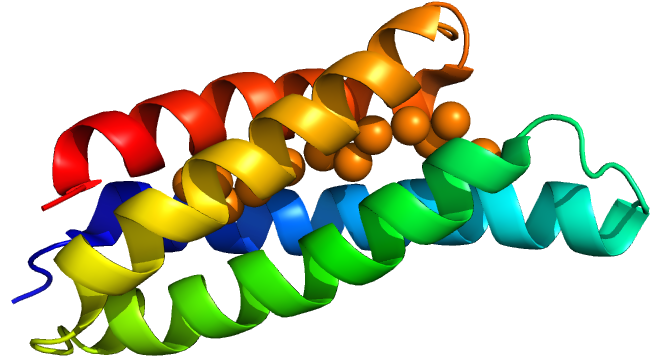 " width="50%"/>
" width="50%"/>
Experience
We were unable however to detect copper chelation activity of Csp1 in vivo using a BCS absorbance assay. Modelling by our team suggested that this was because insufficient protein could be expressed to chelate the amount (units of μmols) needed to be detectable on the assay.
Using a version of this part with a C-terminal His tagged-sfGFP we purified this protein but were still unable to detect copper chelation with the assay in our purified extracts.
