Difference between revisions of "Part:BBa K2110105"
(→Results) |
|||
| Line 12: | Line 12: | ||
<partinfo>BBa_K2110105 SequenceAndFeatures</partinfo> | <partinfo>BBa_K2110105 SequenceAndFeatures</partinfo> | ||
===Results=== | ===Results=== | ||
| + | |||
| + | <h2>Assay system—CFPS system</h2> | ||
| + | <h3>CFPS system</h3> | ||
| + | Cell-free protein synthesis(CFPS) is a widely used method in molecular biology. Production of proteins using cell-free protein synthesis usually takes a few hours, in contrast to production of proteins in cells, which typically takes days to weeks. In fact, even first-time users can often obtain newly synthesized proteins in one day using a commercial system. | ||
| + | |||
| + | The diversity of the cell-free systems allows in vitro synthesis of a wide range of proteins for a variety of downstream applications, such as screeening of enzymes activities. In the post-genomic era, cell-free protein synthesis has rapidly become the preferred approach for high-throughput functional and structural studies of proteins and a versatile tool for in vitro protein evolution and synthetic biology.[1] | ||
| + | |||
| + | <h3>Plasmid construction and expression in CFPS system</h3> | ||
| + | Ligaion of digested pRset_CFP-1, digested CFP gene and digested PETase(I208V) gene in accordance with the 1:5:5 molecular ratio. The newly constructed plasmid is called pRset_CFP-1_PETase-I208V. | ||
| + | Than , the plasmids pRset_CFP-1_PETase-I208V was put into the CFPS(Cell-Free Protein Synthesis) to synthesis the enzymes we expected. | ||
| + | |||
| + | https://static.igem.org/mediawiki/2016/e/e6/T--Tianjin--part-I208Vpc.png | ||
| + | |||
| + | <h3>Dgradation data</h3> | ||
| + | After the enzymes were expressed in the system successfully, we used the proteins we got to degrade PET and detected the degradation product, MHET, which has no other characteristic adsorption peak except in 260nm. | ||
| + | |||
We detect the absorption peak of culture medium after culturing with PET film added for 5d. The absorption degree-wavelength curve is as follows. | We detect the absorption peak of culture medium after culturing with PET film added for 5d. The absorption degree-wavelength curve is as follows. | ||
https://static.igem.org/mediawiki/parts/thumb/7/7a/T--Tianjin--partdataI208V.png/800px-T--Tianjin--partdataI208V.png | https://static.igem.org/mediawiki/parts/thumb/7/7a/T--Tianjin--partdataI208V.png/800px-T--Tianjin--partdataI208V.png | ||
| Line 18: | Line 34: | ||
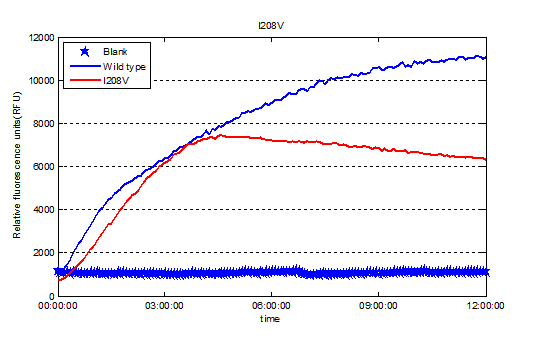
We used the relative fluorescence units to represent the expression level of enzymes. | We used the relative fluorescence units to represent the expression level of enzymes. | ||
| − | + | https://static.igem.org/mediawiki/2016/1/1d/T--Tianjin--part-I208Vex.png | |
Finally we got the relative activities of enzymes. | Finally we got the relative activities of enzymes. | ||
| − | + | https://static.igem.org/mediawiki/2016/2/2f/T--Tianjin--part-I208Vbar.png | |
| + | |||
| + | <h2>Reference</h2> | ||
| + | [1]Shaorong Chong. Overview of Cell-Free Protein Synthesis: Historic Landmarks, Commercial Systems, and Expanding Applications. Current Protocols in Molecular Biology 16.30.1-16.30.11, October 2014 DOI: 10.1002/0471142727.mb1630s108 | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Revision as of 13:50, 15 October 2016
PETase site-directed mutant I208V
This is one of the site-directed mutant of the PETase gene, we change the 208th amino acid from I to V. The module is the Saccharomyces cerevisiae codon optimization version of PETase gene. We use overlap PCR to obtain this mutant.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 79
Illegal BglII site found at 289 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Results
Assay system—CFPS system
CFPS system
Cell-free protein synthesis(CFPS) is a widely used method in molecular biology. Production of proteins using cell-free protein synthesis usually takes a few hours, in contrast to production of proteins in cells, which typically takes days to weeks. In fact, even first-time users can often obtain newly synthesized proteins in one day using a commercial system.
The diversity of the cell-free systems allows in vitro synthesis of a wide range of proteins for a variety of downstream applications, such as screeening of enzymes activities. In the post-genomic era, cell-free protein synthesis has rapidly become the preferred approach for high-throughput functional and structural studies of proteins and a versatile tool for in vitro protein evolution and synthetic biology.[1]
Plasmid construction and expression in CFPS system
Ligaion of digested pRset_CFP-1, digested CFP gene and digested PETase(I208V) gene in accordance with the 1:5:5 molecular ratio. The newly constructed plasmid is called pRset_CFP-1_PETase-I208V. Than , the plasmids pRset_CFP-1_PETase-I208V was put into the CFPS(Cell-Free Protein Synthesis) to synthesis the enzymes we expected.
Dgradation data
After the enzymes were expressed in the system successfully, we used the proteins we got to degrade PET and detected the degradation product, MHET, which has no other characteristic adsorption peak except in 260nm.
We detect the absorption peak of culture medium after culturing with PET film added for 5d. The absorption degree-wavelength curve is as follows.

We used the relative fluorescence units to represent the expression level of enzymes.
Finally we got the relative activities of enzymes.
Reference
[1]Shaorong Chong. Overview of Cell-Free Protein Synthesis: Historic Landmarks, Commercial Systems, and Expanding Applications. Current Protocols in Molecular Biology 16.30.1-16.30.11, October 2014 DOI: 10.1002/0471142727.mb1630s108