Difference between revisions of "Part:BBa K1732000"
| (21 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
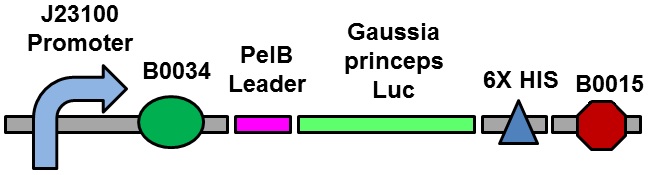
J23100 Promoter with a RBS (BBa_S04423), PelB Leader sequence, Gaussia princeps luciferase, and a B0015 terminator. Allows Gaussia princeps luciferase to be transcribed and translated using a strong promoter. Luminescence can be expressed by reacting the luciferase product with coelenterazine. | J23100 Promoter with a RBS (BBa_S04423), PelB Leader sequence, Gaussia princeps luciferase, and a B0015 terminator. Allows Gaussia princeps luciferase to be transcribed and translated using a strong promoter. Luminescence can be expressed by reacting the luciferase product with coelenterazine. | ||
| + | |||
| + | ---- | ||
| + | |||
| + | [[File:Donnaaddsequencehere.jpg]] | ||
| + | |||
| + | ''Gaussia'' Luciferase (GLuc) is a naturally secreted protein from ''Gaussia princeps''. It is considered the brightest luciferase and its size is around 19kDa. This luciferase is stable at high temperatures due to the presence of disulfide bonds in its structure. This stability makes it an ideal reporter gene, specifically for coelenterazine [1]. | ||
| + | |||
| + | '''Bioluminescent Mechanism of Coelenterazine''' | ||
| + | |||
| + | [[File:Example23.jpg]] | ||
| + | [2] | ||
| + | |||
| + | Coelenterazine is found in several aquatic organisms and is a luminescent substrate for many luciferase enzymes. For this study, PelB Gaussia Luciferase interacted with coelenterazine to determine relative light output and kinetics [3]. | ||
| + | |||
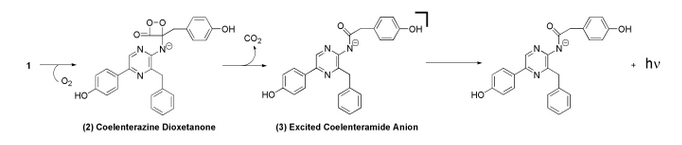
| + | In the mechanism, coelenterazine reacts with oxygen to yield 1,2-dioxetane (compound 2). Subsequent loss of carbon dioxide leads to the intermediate, followed by emission of a photon [2]. | ||
| + | |||
| + | The image above is the chemiluminescent mechanism of coelenterazine. The bioluminescent mechanism is similar, except that the excited state molecule in bioluminescent is phenolate anion instead of the amide anion [2]. | ||
| + | |||
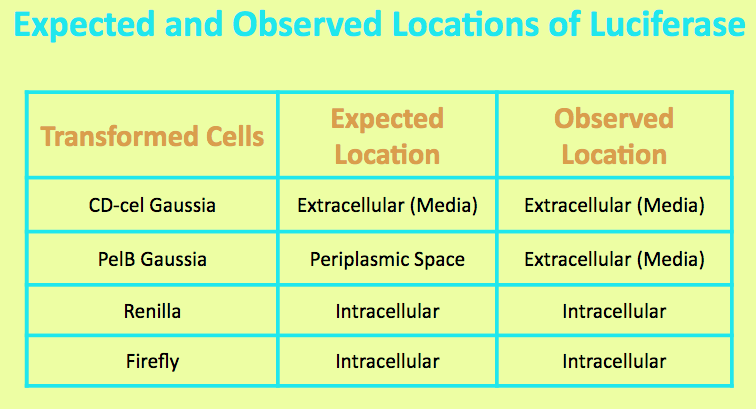
| + | Before running experiments with Gaussia Luciferase, the location of the luciferase in the media was determined by comparing the light output of overnight grown culture of CD-cel Gaussia cells with those of the pellet and the supernatant. The PelB Gaussia culture was spun down, and the pellet represented intracellular localization while the supernatant represented extracellular localization. | ||
| + | |||
| + | |||
| + | [[File:location.jpg]] | ||
| + | |||
| + | |||
| + | It was expected that the CD-cel Luciferase was localized in the cell and the level of light output from the pellet matched that of the overnight culture and was significantly higher than that of the media. This suggested that the luciferase is located intracellularly. | ||
| + | |||
| + | |||
| + | [[File:PelBConc.jpg]] | ||
| + | |||
| + | |||
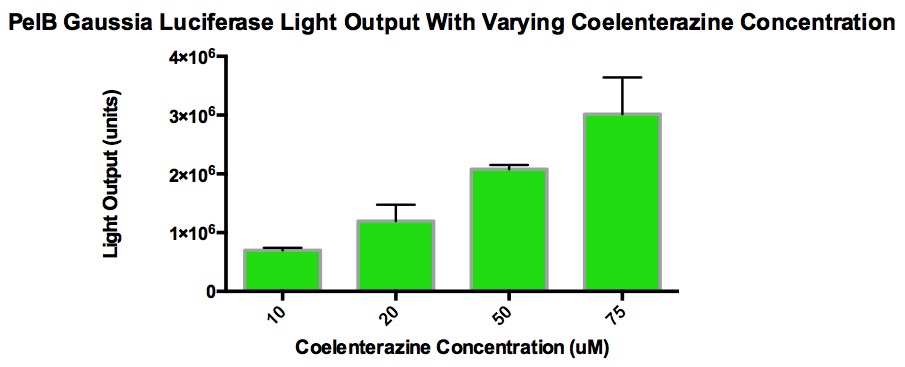
| + | The graph shows the amount of light output produced from varying concentrations of coelenterazine when added to 100uL of PelB Gaussia cells grown overnight (5mL LB broth, 5uL Chloramphenicol, single cell colony). 10uL of coelenterazine was added and this volume stayed consistent. The overnight culture was diluted 1/10 with LB broth in order to measure the Klett OD and stay within accurate measurement range. The purpose of this is to see the effect of light output with increasing concentrations of coelenterazine. The goal was to find a concentration that plateau without maxing out the luminometer (TECAN). It was discovered that at a certain concentration, the light output stop increasing because the acidic methanol used to make the stock solution of coelenterazine began to interfere with the enzyme activity. Also, CD-cel luciferase stays in the cell, which hinders the reaction with coelenterazine. This explains why high concentrations of coelenterazine produces relatively low amounts of light. | ||
| + | |||
| + | |||
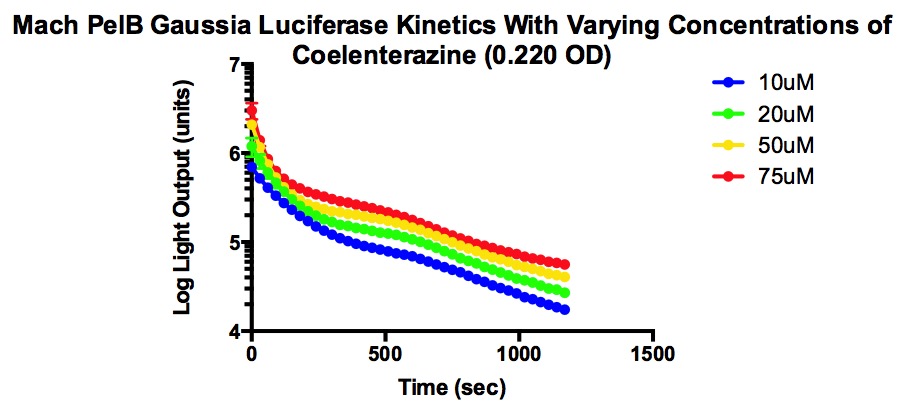
| + | [[File:PelbKinetics.jpg]] | ||
| + | |||
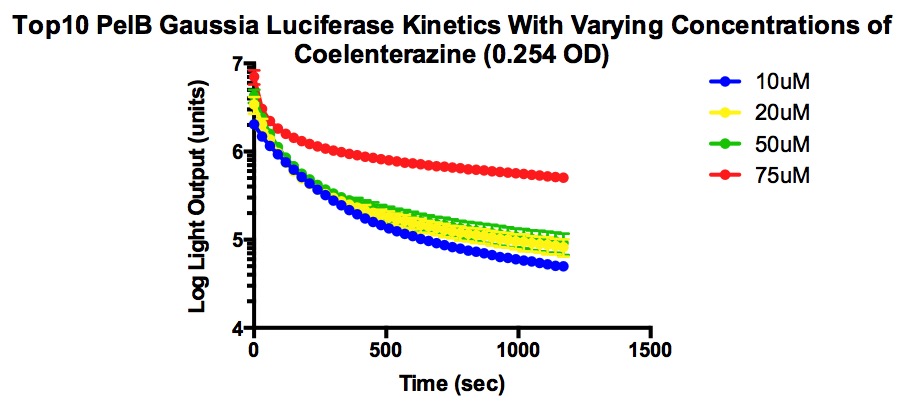
| + | [[File:Top10Pelb.jpg]] | ||
| + | |||
| + | |||
| + | The two types of competent cells used were Mach and Top10 cells. Mach cells are one of the fastest growing competent strain and Top10 cells also have transformation and cloning efficiency. Both are able to replicate high number of plasmids at stable levels. | ||
| + | |||
| + | From the graphs, it can be determined that higher concentrations of coelenterazine allows for a higher light output, but the decay rate is more significant . In addition, the plateau level steadies at a higher light output when given higher concentrations of coelenterazine. It is also important to note that the plateau occurs quickly and the jump in light output is temporary. | ||
| + | |||
| + | |||
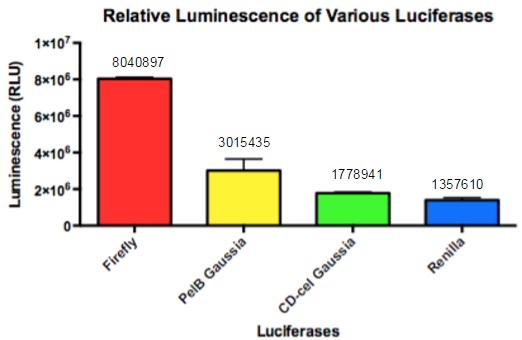
| + | [[File:DONNA3.jpg]] | ||
| + | |||
| + | |||
| + | [[File:Registry2.jpg]] | ||
| + | |||
| + | |||
| + | '''References''' | ||
| + | |||
| + | [1]Gaussia Luciferase [Fact sheet]. (n.d.). Retrieved September 14, 2015, from New England BioLabs Inc. | ||
| + | website: https://www.neb.com/applications/cellular-analysis/reporter-systems/gaussia-luciferase | ||
| + | [2]Gonzalez, V. M., Jr. (2007). Synthesis, Luminescence, and Applications of Coelenterazine and its | ||
| + | Analogs. University of Illinois Urbana-Champaign. | ||
| + | [3]Coelenterazine [Fact sheet]. (n.d.). Retrieved September 14, 2015, from Gold Biotechnology website: | ||
| + | https://www.goldbio.com/product/1012/coelenterazine | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 17: | Line 74: | ||
<partinfo>BBa_K1732000 parameters</partinfo> | <partinfo>BBa_K1732000 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 03:45, 19 November 2015
J23100-PelB-GlucHCO-B0015
J23100 Promoter with a RBS (BBa_S04423), PelB Leader sequence, Gaussia princeps luciferase, and a B0015 terminator. Allows Gaussia princeps luciferase to be transcribed and translated using a strong promoter. Luminescence can be expressed by reacting the luciferase product with coelenterazine.
Gaussia Luciferase (GLuc) is a naturally secreted protein from Gaussia princeps. It is considered the brightest luciferase and its size is around 19kDa. This luciferase is stable at high temperatures due to the presence of disulfide bonds in its structure. This stability makes it an ideal reporter gene, specifically for coelenterazine [1].
Bioluminescent Mechanism of Coelenterazine
Coelenterazine is found in several aquatic organisms and is a luminescent substrate for many luciferase enzymes. For this study, PelB Gaussia Luciferase interacted with coelenterazine to determine relative light output and kinetics [3].
In the mechanism, coelenterazine reacts with oxygen to yield 1,2-dioxetane (compound 2). Subsequent loss of carbon dioxide leads to the intermediate, followed by emission of a photon [2].
The image above is the chemiluminescent mechanism of coelenterazine. The bioluminescent mechanism is similar, except that the excited state molecule in bioluminescent is phenolate anion instead of the amide anion [2].
Before running experiments with Gaussia Luciferase, the location of the luciferase in the media was determined by comparing the light output of overnight grown culture of CD-cel Gaussia cells with those of the pellet and the supernatant. The PelB Gaussia culture was spun down, and the pellet represented intracellular localization while the supernatant represented extracellular localization.
It was expected that the CD-cel Luciferase was localized in the cell and the level of light output from the pellet matched that of the overnight culture and was significantly higher than that of the media. This suggested that the luciferase is located intracellularly.
The graph shows the amount of light output produced from varying concentrations of coelenterazine when added to 100uL of PelB Gaussia cells grown overnight (5mL LB broth, 5uL Chloramphenicol, single cell colony). 10uL of coelenterazine was added and this volume stayed consistent. The overnight culture was diluted 1/10 with LB broth in order to measure the Klett OD and stay within accurate measurement range. The purpose of this is to see the effect of light output with increasing concentrations of coelenterazine. The goal was to find a concentration that plateau without maxing out the luminometer (TECAN). It was discovered that at a certain concentration, the light output stop increasing because the acidic methanol used to make the stock solution of coelenterazine began to interfere with the enzyme activity. Also, CD-cel luciferase stays in the cell, which hinders the reaction with coelenterazine. This explains why high concentrations of coelenterazine produces relatively low amounts of light.
The two types of competent cells used were Mach and Top10 cells. Mach cells are one of the fastest growing competent strain and Top10 cells also have transformation and cloning efficiency. Both are able to replicate high number of plasmids at stable levels.
From the graphs, it can be determined that higher concentrations of coelenterazine allows for a higher light output, but the decay rate is more significant . In addition, the plateau level steadies at a higher light output when given higher concentrations of coelenterazine. It is also important to note that the plateau occurs quickly and the jump in light output is temporary.
References
[1]Gaussia Luciferase [Fact sheet]. (n.d.). Retrieved September 14, 2015, from New England BioLabs Inc.
website: https://www.neb.com/applications/cellular-analysis/reporter-systems/gaussia-luciferase
[2]Gonzalez, V. M., Jr. (2007). Synthesis, Luminescence, and Applications of Coelenterazine and its
Analogs. University of Illinois Urbana-Champaign.
[3]Coelenterazine [Fact sheet]. (n.d.). Retrieved September 14, 2015, from Gold Biotechnology website:
https://www.goldbio.com/product/1012/coelenterazine
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 546
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 115
- 1000COMPATIBLE WITH RFC[1000]