Difference between revisions of "Part:BBa K914003"
(→Characterization of pRha) |
|||
| Line 42: | Line 42: | ||
As we can see from the graph above, pRha promoter works as expected and it could be well tuned by concentration of L-rhamnose. | As we can see from the graph above, pRha promoter works as expected and it could be well tuned by concentration of L-rhamnose. | ||
| + | |||
| + | |||
| + | |||
| + | The UChicago 2015 iGEM team also used this part in their project and did some characterization of the promoter, using Western blot for their gene of interest, providing an improvement on the characterization accomplished by the 2012 Paris-Bettencourt team which used a plate-reader to measure fluorescence. In contrast to the low copy plasmid used by the Paris Bettencourt team, they characterized the promoter when expressed in a high copy plasmid. Accordingly, they discovered that despite following a similar protocol (10 hours of induction), significantly less rhamnose was required for the induction of the promoter in the high copy number plasmid. | ||
| + | |||
| + | [[Image:UChicago2015Image002.png|thumb|530px|center|<font size="1">Induction for the characterization curve was accomplished in M9 minimal media with supplements (0.4%glycerol, 0.1% casamino acids).</font>]] | ||
| + | |||
| + | The UChicago team also verified that the pRha promoter is quite tight and has a relatively gradual response curve, characteristics that makes the promoter especially well-suited for assaying a wide range of concentrations of a protein of interest. | ||
| + | |||
| + | The densitometry results from Western blots indicate the tightness of pRha. At 0% L-rhamnose, there is significantly less KaiA expressed compared to KaiB and KaiC on a separate constitutively expressed promoter. This supports our ability to tune KaiA concentrations to optimize oscillations. | ||
| + | |||
| + | [[Image:Image003.jpg|thumb|530px|center|<font size="1">Fitted exponential curve plotted in Prism to %Rhamnose vs. ng of KaiA gradient curve. </font>]] | ||
| + | |||
| + | |||
| + | The gradient curve developed portrays the ability to fine tune the concentrations of KaiA. The data in this graph was corrected using ng from protein standards (below) | ||
| + | [[Image:Image004.jpg|thumb|530px|center|<font size="1">Fitted standard curve of purified Kai A proteins to determine ng of protein from intensity readings in Image J. </font>]] | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 06:23, 25 September 2015
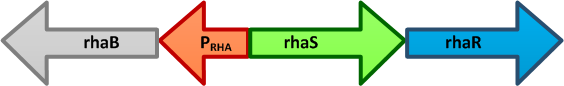
L-rhamnose-inducible promoter (pRha)
L-rhamnose-inducible promoter is capable of high-level recombinant protein expression in the presence of L-rhamnose, it is also tightly regulated in the absence of L-rhamnose by the addition of D-glucose. Read about this biobrick part [http://2012.igem.org/Team:Paris_Bettencourt/Restriction_Enzyme in the context of the project].
Biology of pRha
L-rhamnose is taken up by the RhaT transport system, converted to L-rhamnulose by an isomerase RhaA and then phosphorylated by a kinase RhaB. Subsequently, the resulting rhamnulose-1-phosphate is hydrolyzed by an aldolase RhaD into dihydroxyacetone phosphate, which is metabolized in glycolysis, and L-lactaldehyde. The latter can be oxidized into lactate under aerobic conditions and be reduced into L-1,2-propanediol under unaerobic conditions.
The genes rhaBAD are organized in one operon which is controlled by the rhaPBAD promoter. This promoter is regulated by two activators, RhaS and RhaR, and the corresponding genes belong to one transcription unit which is located in opposite direction of rhaBAD. If L-rhamnose is available, RhaR binds to the rhaPRS promoter and activates the production of RhaR and RhaS. RhaS together with L-rhamnose in turn binds to the rhaPBAD and the rhaPT promoter and activates the transcription of the structural genes. However, for the application of the rhamnose expression system it is not necessary to express the regulatory proteins in larger quantities, because the amounts expressed from the chromosome are sufficient to activate transcription even on multi-copy plasmids. Therefore, only the rhaPBAD promoter has to be cloned upstream of the gene that is to be expressed. Full induction of rhaBAD transcription also requires binding of the CRP-cAMP complex, which is a key regulator of catabolite repression.
The pRha sequence was containing an EcoRI restriction site, so we had to disrupt it in order to use pRha as a biobrick. In order to decide which base pair to modify, we used the [http://microbes.ucsc.edu UCSC Microbial Genome Browser]. We compared the pRha sequence in E.coli and similar species, and identified that the at the position is sometimes replaced by a in some species, so we decided to replace it in a same way. We ordered a gBlock with the pRha sequence having the mutation, and this is the sequence we used and submitted.
Characterization of pRha
Experimental setup
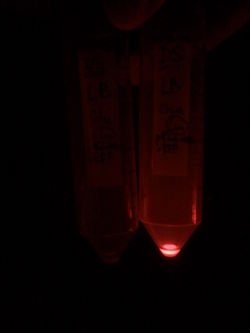
In order to characterize this promoter, we made a construct with a medium RBS (B0032) and an RFP cloned downstream of the pRha, on the pSB3C5 plasmid. We induced the expression of RFP by adding L-Rhamnose. As a negative control, we used cells without the inducer, as well as cells repressed with Glucose.
Results
First, by simple observation under a fluorescence viewer, we have seen that the addition of 1% L-Rhamnose leads to a significant expression of RFP after 10hours. The negative controls, where no Rhamnose was added, or when the promoter was repressed by Glucose, did not show any visible fluorescence. Both photos are taken after we centrifuged a culture of NEB Turbo strain with transformed plasmid. For the fluorescent result, the same tubes were photographed under excitation light (540nm), through an emission filter (590nm).
We quantified this result in a plate reader.
Next, we characterized the pRha promoter using a plate reader. We used different concentrations of L-Rhamnose (0.05%, 0.1%, 0.2%, 0.5% and 1%) and observed the resulting fluorescence over time. As negative controls, we used the non-induced cells, as well as cells repressed by 1% Glucose.
As we can see from the graph above, pRha promoter works as expected and it could be well tuned by concentration of L-rhamnose.
The UChicago 2015 iGEM team also used this part in their project and did some characterization of the promoter, using Western blot for their gene of interest, providing an improvement on the characterization accomplished by the 2012 Paris-Bettencourt team which used a plate-reader to measure fluorescence. In contrast to the low copy plasmid used by the Paris Bettencourt team, they characterized the promoter when expressed in a high copy plasmid. Accordingly, they discovered that despite following a similar protocol (10 hours of induction), significantly less rhamnose was required for the induction of the promoter in the high copy number plasmid.
The UChicago team also verified that the pRha promoter is quite tight and has a relatively gradual response curve, characteristics that makes the promoter especially well-suited for assaying a wide range of concentrations of a protein of interest.
The densitometry results from Western blots indicate the tightness of pRha. At 0% L-rhamnose, there is significantly less KaiA expressed compared to KaiB and KaiC on a separate constitutively expressed promoter. This supports our ability to tune KaiA concentrations to optimize oscillations.
The gradient curve developed portrays the ability to fine tune the concentrations of KaiA. The data in this graph was corrected using ng from protein standards (below)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]