Difference between revisions of "Part:BBa K1639008"
(→References) |
|||
| Line 4: | Line 4: | ||
TEV protease (also called Tobacco Etch Virus nuclear inclusion a endopeptidase) is a highly sequence-specific cysteine protease from Tobacco Etch Virus (TEV).The consensus for these native cut sites is '''ENLYFQ\S''' where ‘\’ denotes the cleaved peptide bond.One of the main uses of this protein is for removing affinity tags from purified proteins. The reason for the use of TEV protease as a biochemical tool is its '''high sequence specificity'''. This specificity allows for the controlled cleavage of proteins when the preference sequence is inserted into flexible loops. It also makes it relatively non-toxic in vivo as the recognized sequence scarcely occurs in proteins[1]. However, TEV protease does have limitations as a biochemical tool. It is prone to deactivation by self-cleavage (autolysis), though this can be abolished through a single S219V mutation in the internal cleavage site.[2] | TEV protease (also called Tobacco Etch Virus nuclear inclusion a endopeptidase) is a highly sequence-specific cysteine protease from Tobacco Etch Virus (TEV).The consensus for these native cut sites is '''ENLYFQ\S''' where ‘\’ denotes the cleaved peptide bond.One of the main uses of this protein is for removing affinity tags from purified proteins. The reason for the use of TEV protease as a biochemical tool is its '''high sequence specificity'''. This specificity allows for the controlled cleavage of proteins when the preference sequence is inserted into flexible loops. It also makes it relatively non-toxic in vivo as the recognized sequence scarcely occurs in proteins[1]. However, TEV protease does have limitations as a biochemical tool. It is prone to deactivation by self-cleavage (autolysis), though this can be abolished through a single S219V mutation in the internal cleavage site.[2] | ||
| − | |||
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | < | + | |
| + | ===Characterization=== | ||
| + | |||
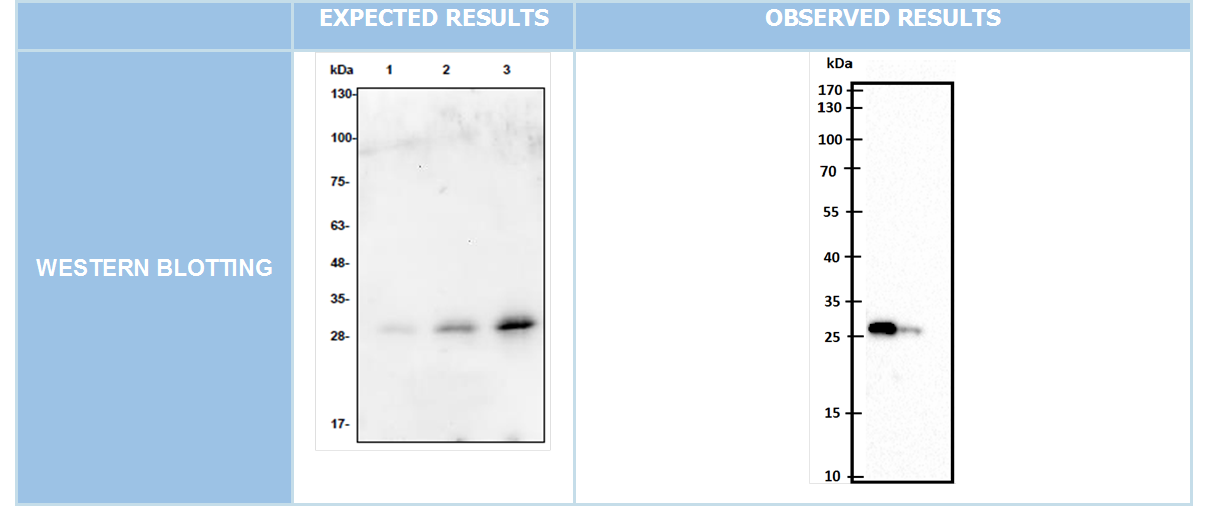
| + | '''WESTERN BLOTTING''' | ||
| + | |||
| + | After we clonned to pET45-b succesfully we showed our aim proteins production via western blotting with using His-tag which locate in N-terminal of these proteins. | ||
| + | |||
| + | [[File:ATOMS-Turkiye_ulcer_damp_5.4.png|600px|thumb|center|<center>'''Figure 2'''</center>]] | ||
| + | |||
| + | '''pColA-DAMP-PEXIGANAN CLONNING''' | ||
| + | |||
| + | We demonstrated that DAMP-Pexiganan and TEV Protease proteins synthesing succesfully within clonning to pET45-b plasmid. We thought that we should insert both two genes(DAMP-Pexiganan and TEV Protease) at the same time to same bacteria to show TEV Protease breaking linker chain (to releasing active free pexiganan) that is between DAMP and Pexiganan. We have seen with the same origin in two plasmids can not be found in a bacterium simultaneously due to origin incompatibility process. For this reason, we decided to clonning one of these two gene sequences into pet45-b plasmid which have ColE origin and one of the gene sequence clonning into pColA plasmid which have ColA origin which also we designed it. | ||
| + | |||
| + | Before that we achieved to clonning both of two gene sequences into pET45-b vector. Therefore, clonning only one of these genes would be enough for us. We decided to locate TEV Protease gene sequence into pET45-b because our essential want to control protein is TEV Protease. And we decided clonning DAMP-Pexiganan gene sequence into pColA vector. For clonning DAMP-Pexiganan protein sequence into pColA vector we toke out our gene with NotI enzyme from PSB1C3-DAMP-Pexiganan plasmid. We did ligation with pET45-b vector which digested the same enzyme. Following ligation, the resulting plasmids were transformed into BL21 competent bacteria. | ||
| + | |||
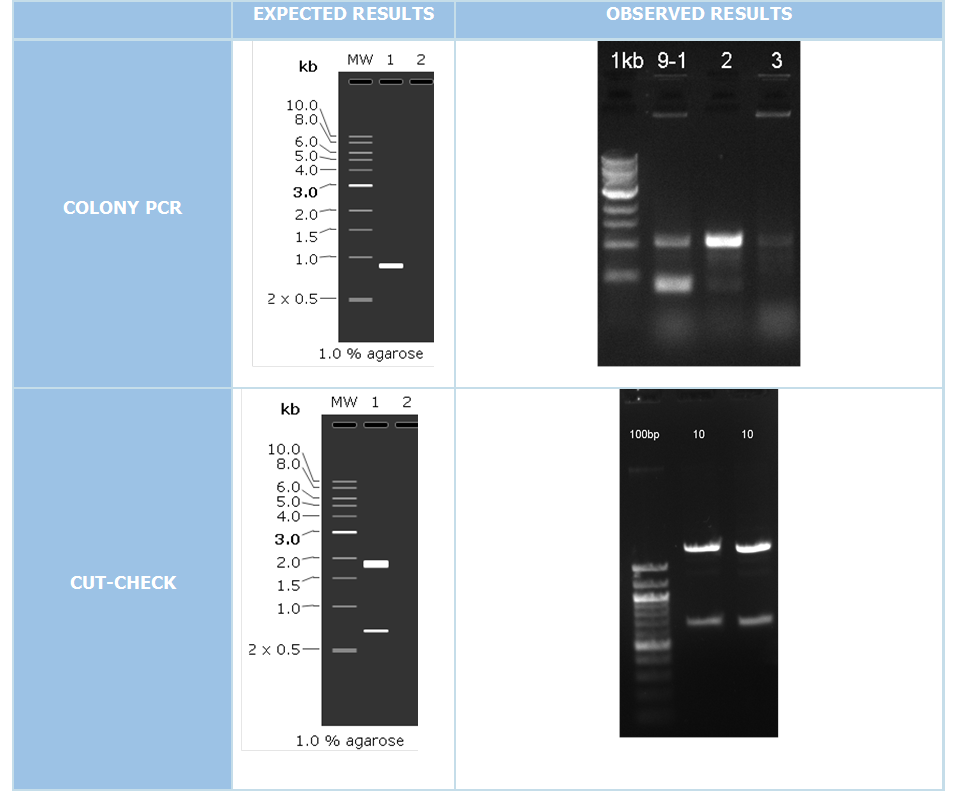
| + | We did colony PCR in order to check the accuracy of cloning with using pColA forward and pColA reverse primers. If the clonning resulted negative expecting band would be 192 bp. On the other hand if we succesfully clonned these part, expected band would be 908 bp. | ||
| + | |||
| + | After colony PCR consequences we have put the positive resulted clonnings 16 hours liquid culture after that we isolated plasmid DNA with miniprep plasmid izolation method. We make cutcheck to our attained plasmid DNAs for put a second control step. Our restriction enzymes those we used are XbaI and SpeI. | ||
| + | |||
| + | [[File:ATOMS-Turkiye_ulcer_damp_5.5.png|600px|thumb|center|<center>'''Figure 2'''</center>]] | ||
| + | |||
| + | '''pColA- DAMP-PEXIGANAN/pET45-TEV PROTEASE CO-TRANSFORMATION''' | ||
| + | |||
| + | After we clonning DAMP-Pexiganan gene sequence into pColA plasmid and TEV Protease gene sequence into pET45-b for transformate these both two plasmids into same BL21 bacteria we did cotransformation. After cotransformation process we put bacterias at 37C for 16 hours incubation. End of 16 hours as a result we did not see any bacteria colony over agar plate. | ||
| + | |||
| + | We retried several times this cotransformation process but with these tryings we havent seen any bacteria colony on our agar plates. | ||
| + | |||
| + | '''TEV protease Activity Control''' | ||
| + | |||
| + | After the negative results obtained from cotransformation experiements we designed a new experimental apparatus to demonstrate TEV Protease's breaking or separator effect to DAMP-Pexiganan molecules linker site. We firstly make broth culture secondly put incubation at 37C for 13 hours bacterias which contain pET45-DAMP-Pexiganan and pET45-TEV Protease plasmids. For inducing protein production we added 1mM IPTG into medium at 13rd hour. Next we left these incubation for 3 hours at 37C. We isolated protein from totally 16 hours incubated broth culture. We prepared a suitable buffer to create the conditions necessary to show the TEV Protease’s enzymatic activity.(Conditions are given below). Then we prepare our DAMP-Pex, and TEV protease isolated proteins by mixing in the buffer, we put the resulting mixture overnight incubation at 20° C. After incubation, to observe the fate of DAMP-pexigan proteins we did Western Blot. But in western blotting, we could not get any significant rational result. | ||
| + | |||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_K1639008 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1639008 SequenceAndFeatures</partinfo> | ||
| + | |||
===References=== | ===References=== | ||
[1]Parks TD, Leuther KK, Howard ED, Johnston SA, Dougherty WG (February 1994). "Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase". Anal. Biochem. 216 (2): 413–7. doi:10.1006/abio.1994.1060. PMID 8179197 | [1]Parks TD, Leuther KK, Howard ED, Johnston SA, Dougherty WG (February 1994). "Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase". Anal. Biochem. 216 (2): 413–7. doi:10.1006/abio.1994.1060. PMID 8179197 | ||
| − | + | <br>[2]Kapust RB, Tözsér J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS (December 2001). "Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency". Protein Eng. 14 (12): 993–1000. doi:10.1093/protein/14.12.993. PMID 11809930 | |
| − | [2]Kapust RB, Tözsér J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS (December 2001). "Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency". Protein Eng. 14 (12): 993–1000. doi:10.1093/protein/14.12.993. PMID 11809930 | + | |
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Revision as of 13:14, 21 September 2015
Tev Protease, single S219V mutation in the internal cleavage site
TEV protease (also called Tobacco Etch Virus nuclear inclusion a endopeptidase) is a highly sequence-specific cysteine protease from Tobacco Etch Virus (TEV).The consensus for these native cut sites is ENLYFQ\S where ‘\’ denotes the cleaved peptide bond.One of the main uses of this protein is for removing affinity tags from purified proteins. The reason for the use of TEV protease as a biochemical tool is its high sequence specificity. This specificity allows for the controlled cleavage of proteins when the preference sequence is inserted into flexible loops. It also makes it relatively non-toxic in vivo as the recognized sequence scarcely occurs in proteins[1]. However, TEV protease does have limitations as a biochemical tool. It is prone to deactivation by self-cleavage (autolysis), though this can be abolished through a single S219V mutation in the internal cleavage site.[2]
Usage and Biology
Characterization
WESTERN BLOTTING
After we clonned to pET45-b succesfully we showed our aim proteins production via western blotting with using His-tag which locate in N-terminal of these proteins.
pColA-DAMP-PEXIGANAN CLONNING
We demonstrated that DAMP-Pexiganan and TEV Protease proteins synthesing succesfully within clonning to pET45-b plasmid. We thought that we should insert both two genes(DAMP-Pexiganan and TEV Protease) at the same time to same bacteria to show TEV Protease breaking linker chain (to releasing active free pexiganan) that is between DAMP and Pexiganan. We have seen with the same origin in two plasmids can not be found in a bacterium simultaneously due to origin incompatibility process. For this reason, we decided to clonning one of these two gene sequences into pet45-b plasmid which have ColE origin and one of the gene sequence clonning into pColA plasmid which have ColA origin which also we designed it.
Before that we achieved to clonning both of two gene sequences into pET45-b vector. Therefore, clonning only one of these genes would be enough for us. We decided to locate TEV Protease gene sequence into pET45-b because our essential want to control protein is TEV Protease. And we decided clonning DAMP-Pexiganan gene sequence into pColA vector. For clonning DAMP-Pexiganan protein sequence into pColA vector we toke out our gene with NotI enzyme from PSB1C3-DAMP-Pexiganan plasmid. We did ligation with pET45-b vector which digested the same enzyme. Following ligation, the resulting plasmids were transformed into BL21 competent bacteria.
We did colony PCR in order to check the accuracy of cloning with using pColA forward and pColA reverse primers. If the clonning resulted negative expecting band would be 192 bp. On the other hand if we succesfully clonned these part, expected band would be 908 bp.
After colony PCR consequences we have put the positive resulted clonnings 16 hours liquid culture after that we isolated plasmid DNA with miniprep plasmid izolation method. We make cutcheck to our attained plasmid DNAs for put a second control step. Our restriction enzymes those we used are XbaI and SpeI.
pColA- DAMP-PEXIGANAN/pET45-TEV PROTEASE CO-TRANSFORMATION
After we clonning DAMP-Pexiganan gene sequence into pColA plasmid and TEV Protease gene sequence into pET45-b for transformate these both two plasmids into same BL21 bacteria we did cotransformation. After cotransformation process we put bacterias at 37C for 16 hours incubation. End of 16 hours as a result we did not see any bacteria colony over agar plate.
We retried several times this cotransformation process but with these tryings we havent seen any bacteria colony on our agar plates.
TEV protease Activity Control
After the negative results obtained from cotransformation experiements we designed a new experimental apparatus to demonstrate TEV Protease's breaking or separator effect to DAMP-Pexiganan molecules linker site. We firstly make broth culture secondly put incubation at 37C for 13 hours bacterias which contain pET45-DAMP-Pexiganan and pET45-TEV Protease plasmids. For inducing protein production we added 1mM IPTG into medium at 13rd hour. Next we left these incubation for 3 hours at 37C. We isolated protein from totally 16 hours incubated broth culture. We prepared a suitable buffer to create the conditions necessary to show the TEV Protease’s enzymatic activity.(Conditions are given below). Then we prepare our DAMP-Pex, and TEV protease isolated proteins by mixing in the buffer, we put the resulting mixture overnight incubation at 20° C. After incubation, to observe the fate of DAMP-pexigan proteins we did Western Blot. But in western blotting, we could not get any significant rational result.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 2
Illegal XhoI site found at 744 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 605
References
[1]Parks TD, Leuther KK, Howard ED, Johnston SA, Dougherty WG (February 1994). "Release of proteins and peptides from fusion proteins using a recombinant plant virus proteinase". Anal. Biochem. 216 (2): 413–7. doi:10.1006/abio.1994.1060. PMID 8179197
[2]Kapust RB, Tözsér J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS (December 2001). "Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency". Protein Eng. 14 (12): 993–1000. doi:10.1093/protein/14.12.993. PMID 11809930