Difference between revisions of "Part:BBa K1689008"
| Line 18: | Line 18: | ||
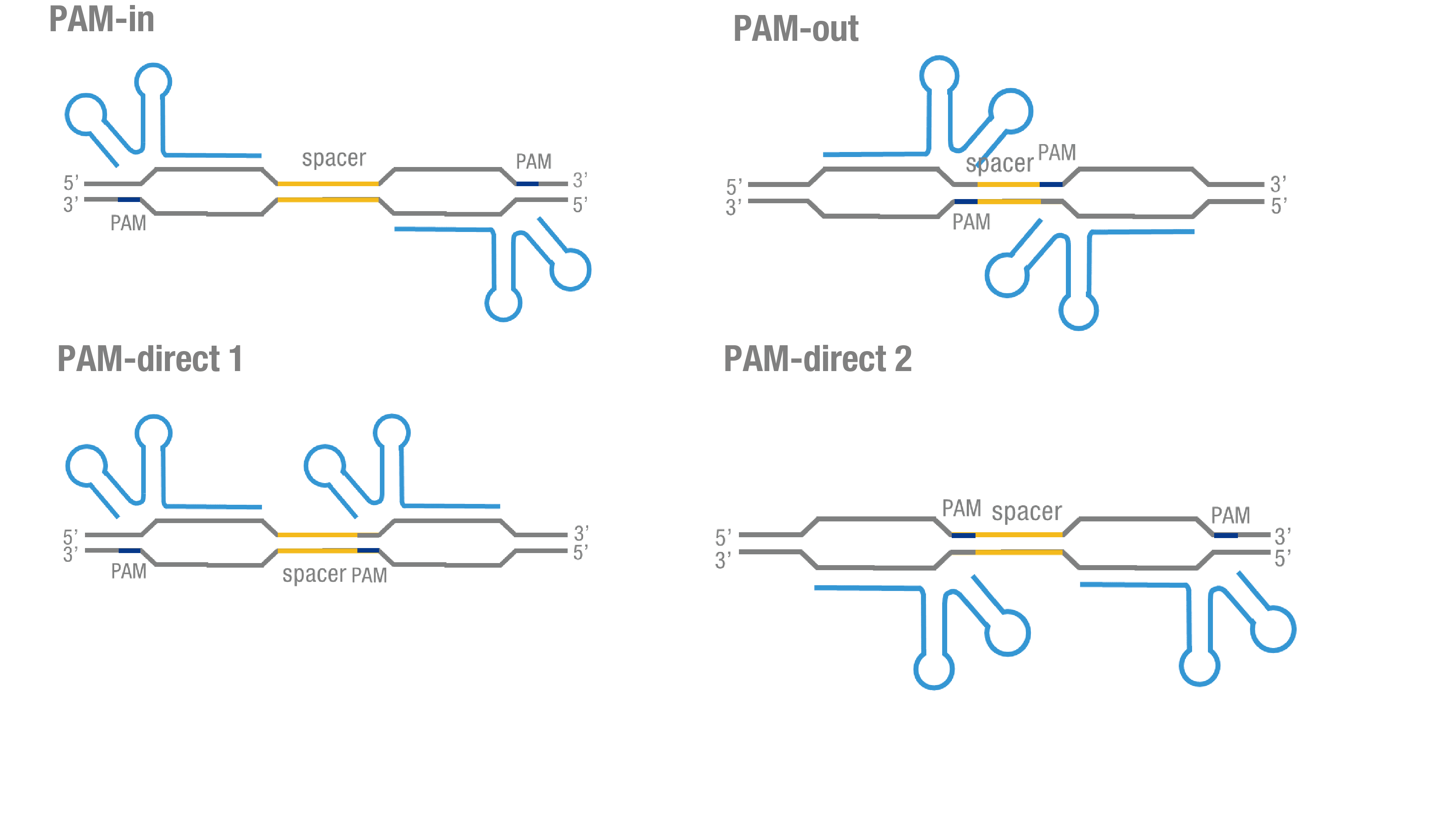
| − | We thoroughly optimized the configuration of our PC reporter system (see [http://2015.igem.org/Team:Peking/Design/PC_Reporter Methods]). Provided that the initial binding of dCas9 to DNA depends on the protospacer adjacent motif (PAM, a short 3’ motif adjacent to target sequence), four sets of sgRNA orientation settings were tested (Figure 3a).To find out how split luciferase-dCas9 fusion strategy influences our PC reporter system, we constructed and tested | + | We thoroughly optimized the configuration of our PC reporter system (see [http://2015.igem.org/Team:Peking/Design/PC_Reporter Methods]). Provided that the initial binding of dCas9 to DNA depends on the protospacer adjacent motif (PAM, a short 3’ motif adjacent to target sequence), four sets of sgRNA orientation settings were tested (Figure 3a).To find out how split luciferase-dCas9 fusion strategy influences our PC reporter system, we constructed and tested C-luc-dCas9[https://parts.igem.org/wiki/index.php?title=Part:BBa_K1689009 (BBa_K1689009)], dCas9-Cluc [https://parts.igem.org/wiki/index.php?title=Part:BBa_K1689007 (BBa_K1689007) ]fusion protein to respectively pair with dCas9-Nluc (Figure 3b). |
a | a | ||
| Line 28: | Line 28: | ||
[[File:Peking-11.png|700px|]] | [[File:Peking-11.png|700px|]] | ||
| − | Figure 3. Thorough optimization on the configuration of our PC reporter system. (a) 4 different sgRNA orientation settings. In orientation PAM-out, the pair of PAM sequences are distal from the spacer sequence, with the 5' end of the sgRNA adjacent to the spacer; in orientation PAM-in, the pair of PAM sequences are adjacent to the spacer sequence, with the 3' end of the sgRNA in proximity to the spacer; in orientation PAM-direct 1 and PAM-direct 2, one PAM sequence is adjacent to and another distal from the spacer. (b) Test on dCas9-Nluc fusion strategies paired with | + | Figure 3. Thorough optimization on the configuration of our PC reporter system. (a) 4 different sgRNA orientation settings. In orientation PAM-out, the pair of PAM sequences are distal from the spacer sequence, with the 5' end of the sgRNA adjacent to the spacer; in orientation PAM-in, the pair of PAM sequences are adjacent to the spacer sequence, with the 3' end of the sgRNA in proximity to the spacer; in orientation PAM-direct 1 and PAM-direct 2, one PAM sequence is adjacent to and another distal from the spacer. (b) Test on dCas9-Nluc fusion strategies paired with C-luc-dCas9 and dCas9-C-luc across four different sgRNA orientations. |
Revision as of 04:06, 19 September 2015
dCas9-N-luc
dCas9-N-luc fusion protein ORF
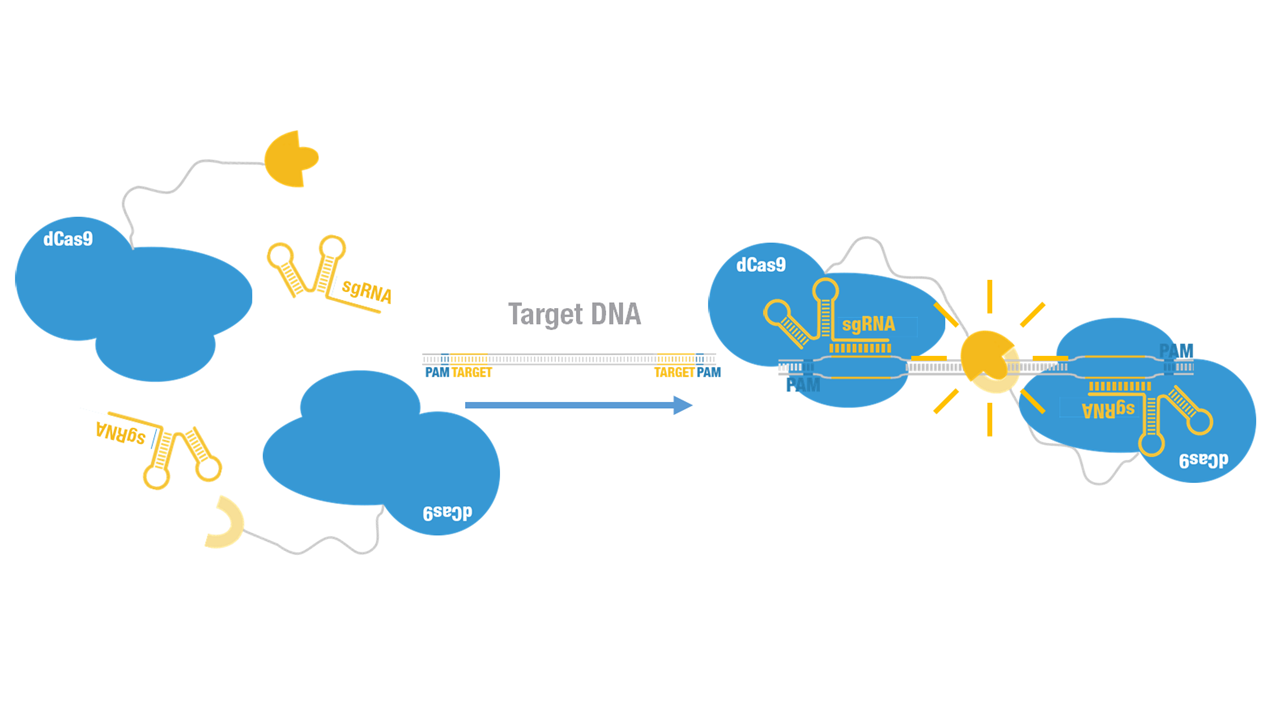
A catalytically dead Cas9 (dCas9), when co-expressed with a guide RNA, forms a DNA recognition complex which can binds any sequence [1]. Firefly luciferase, widely used as a reporter, is split into two fragments, namely N-luc and C-luc [2]. Each fragment by itself is inactive; when two fragments are reassembled, the enzymatic activity of the original protein would be reconstituted, providing easily measurable readout.
2015 Peking iGEM fused N-luc to C terminus of dCas9 (Figure 1). Guided by sgRNA, it binds to target DNA sequence. Together with another part, dCas9-C-luc (BBa_K1689007):sgRNA complex, our paired dCas9 (PC) reporter system would work to (Figure 2) to convert the sequence-specific information of pathogenic bacteria's genome (in our case, M. tuberculosis) into easily readable bioluminescence signal.
Figure 1. Schematic cartoon of dCas9-N-luc fusion protein.
Figure 2. Working mechanism of the paired dCas9 (PC) reporter system.
We thoroughly optimized the configuration of our PC reporter system (see [http://2015.igem.org/Team:Peking/Design/PC_Reporter Methods]). Provided that the initial binding of dCas9 to DNA depends on the protospacer adjacent motif (PAM, a short 3’ motif adjacent to target sequence), four sets of sgRNA orientation settings were tested (Figure 3a).To find out how split luciferase-dCas9 fusion strategy influences our PC reporter system, we constructed and tested C-luc-dCas9(BBa_K1689009), dCas9-Cluc (BBa_K1689007) fusion protein to respectively pair with dCas9-Nluc (Figure 3b).
a
b
Figure 3. Thorough optimization on the configuration of our PC reporter system. (a) 4 different sgRNA orientation settings. In orientation PAM-out, the pair of PAM sequences are distal from the spacer sequence, with the 5' end of the sgRNA adjacent to the spacer; in orientation PAM-in, the pair of PAM sequences are adjacent to the spacer sequence, with the 3' end of the sgRNA in proximity to the spacer; in orientation PAM-direct 1 and PAM-direct 2, one PAM sequence is adjacent to and another distal from the spacer. (b) Test on dCas9-Nluc fusion strategies paired with C-luc-dCas9 and dCas9-C-luc across four different sgRNA orientations.
References
1. Lei S. Qi, Matthew H. Larson, Luke A. Gilbert et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell, 2013, 152: 1173-1183.
2. Kathryn E. Luker, Matthew C. P. Smith, et al. Kinetics of regulated protein–protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. PNAS, 2004, 101: 12288-12293.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1176
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 3
Illegal BamHI site found at 3455 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]