Difference between revisions of "Part:BBa K1761000:Design"
(figures) |
(headings) |
||
| Line 6: | Line 6: | ||
===Design Notes=== | ===Design Notes=== | ||
| + | '''Sequence adoptations''' | ||
| + | Codon optimalization and deleting restriction sites used for biobricking and classical cloning. | ||
| − | + | ===Source=== | |
| + | An outer membrane protein naturally occuring in wildtype E.coli. | ||
| − | |||
| − | + | == The Protein OmpX == | |
| + | ===Outer Membrane Proteins=== | ||
Outer membrane proteins form an essential component for Gram-negative bacteria, providing the bacteria with protection against a harsh environment [1]. More than six outer membrane protein families have been discovered, which all share a beta barrel secondary structure. Thanks to this structure, the proteins feature loops protruding from the bacterial outer membrane, and intracellular C-termini. This enables clicking of aptamers to the membrane proteins and fusing signaling components to the outer membrane proteins intracellularly, making outer membrane proteins the perfect scaffold for COMBs, the iGEM TU Eindhoven 2015 project [http://2015.igem.org/Team:TU_Eindhoven]. | Outer membrane proteins form an essential component for Gram-negative bacteria, providing the bacteria with protection against a harsh environment [1]. More than six outer membrane protein families have been discovered, which all share a beta barrel secondary structure. Thanks to this structure, the proteins feature loops protruding from the bacterial outer membrane, and intracellular C-termini. This enables clicking of aptamers to the membrane proteins and fusing signaling components to the outer membrane proteins intracellularly, making outer membrane proteins the perfect scaffold for COMBs, the iGEM TU Eindhoven 2015 project [http://2015.igem.org/Team:TU_Eindhoven]. | ||
| − | |||
| + | ===The Scaffold - OmpX=== | ||
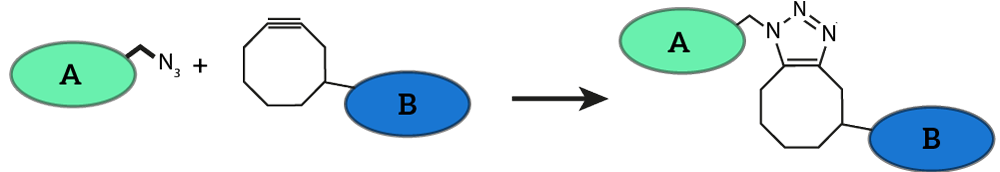
A procedure to attach oligonucleotides to membrane proteins has succesfully been tested by iGEM TU Eindhoven 2014 [http://2014.igem.org/Team:TU_Eindhoven]. They presented this procedure as a part of their Click Coli project to the iGEM community. The procedure exploits the SPAAC reaction, which is one of the most well-described click chemistry reactions available (see Figure 1). | A procedure to attach oligonucleotides to membrane proteins has succesfully been tested by iGEM TU Eindhoven 2014 [http://2014.igem.org/Team:TU_Eindhoven]. They presented this procedure as a part of their Click Coli project to the iGEM community. The procedure exploits the SPAAC reaction, which is one of the most well-described click chemistry reactions available (see Figure 1). | ||
| Line 26: | Line 29: | ||
As a proof of concept, iGEM TU Eindhoven 2014 employed two membrane proteins as anchors, to which they anchored DNA. These membrane proteins were CPX and INPNC. CPX in particular showed promising results and is still being explored today. | As a proof of concept, iGEM TU Eindhoven 2014 employed two membrane proteins as anchors, to which they anchored DNA. These membrane proteins were CPX and INPNC. CPX in particular showed promising results and is still being explored today. | ||
| − | + | ===CPX & OmpX=== | |
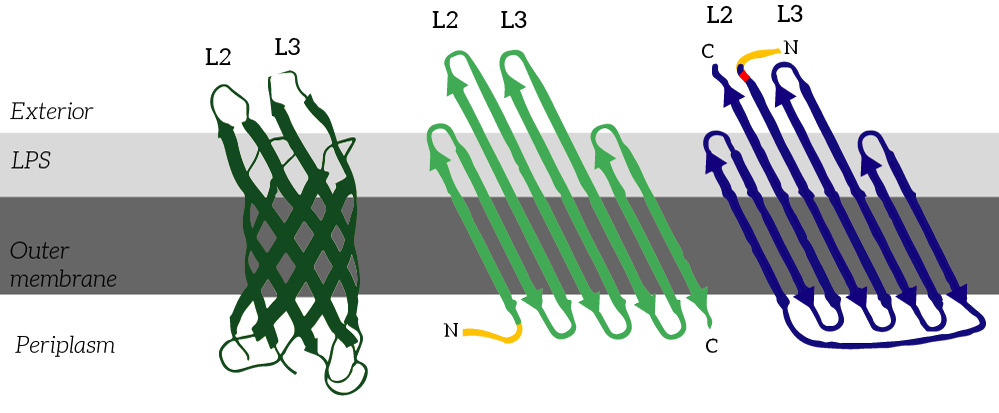
CPX is a membrane display protein developed by Rice et al. as an alternative to traditional display methodologies, such as yeast display and phage display [2]. The protein itself is derived from the naturally occuring Outer membrane protein X (OmpX) through circular permutation (see Figure 2). Circular permutation was a necessary step to obtain the termini on the exterior of the bacterial cells. | CPX is a membrane display protein developed by Rice et al. as an alternative to traditional display methodologies, such as yeast display and phage display [2]. The protein itself is derived from the naturally occuring Outer membrane protein X (OmpX) through circular permutation (see Figure 2). Circular permutation was a necessary step to obtain the termini on the exterior of the bacterial cells. | ||
| Line 49: | Line 52: | ||
| − | + | ===The Loops=== | |
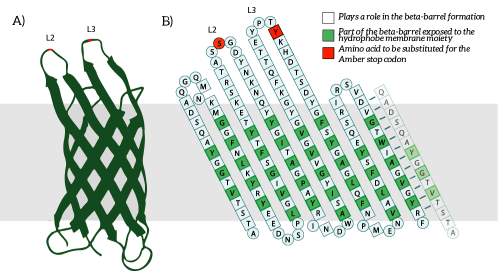
The Amber stop codon will be incorporated in the protruding loops of OmpX. We have chosen to introduce the mutations into the loops since they are easily accessible and are not a part of the beta-barrel of OmpX [2]. Hence, we believe that mutations in the loops will not disturb the secondary structure of OmpX (see Figure 5B). | The Amber stop codon will be incorporated in the protruding loops of OmpX. We have chosen to introduce the mutations into the loops since they are easily accessible and are not a part of the beta-barrel of OmpX [2]. Hence, we believe that mutations in the loops will not disturb the secondary structure of OmpX (see Figure 5B). | ||
| Line 63: | Line 66: | ||
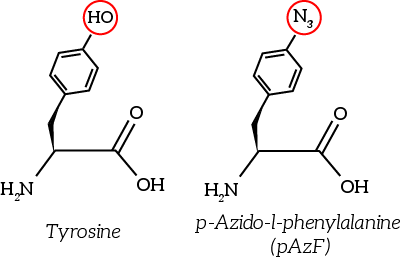
''Figure 6: The non-natural azide-functionalized amino acid closely resembles tyrosine'' | ''Figure 6: The non-natural azide-functionalized amino acid closely resembles tyrosine'' | ||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 16:26, 15 September 2015
Outer Membrane Protein X (OmpX)
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Design Notes
Sequence adoptations Codon optimalization and deleting restriction sites used for biobricking and classical cloning.
Source
An outer membrane protein naturally occuring in wildtype E.coli.
The Protein OmpX
Outer Membrane Proteins
Outer membrane proteins form an essential component for Gram-negative bacteria, providing the bacteria with protection against a harsh environment [1]. More than six outer membrane protein families have been discovered, which all share a beta barrel secondary structure. Thanks to this structure, the proteins feature loops protruding from the bacterial outer membrane, and intracellular C-termini. This enables clicking of aptamers to the membrane proteins and fusing signaling components to the outer membrane proteins intracellularly, making outer membrane proteins the perfect scaffold for COMBs, the iGEM TU Eindhoven 2015 project [http://2015.igem.org/Team:TU_Eindhoven].
The Scaffold - OmpX
A procedure to attach oligonucleotides to membrane proteins has succesfully been tested by iGEM TU Eindhoven 2014 [http://2014.igem.org/Team:TU_Eindhoven]. They presented this procedure as a part of their Click Coli project to the iGEM community. The procedure exploits the SPAAC reaction, which is one of the most well-described click chemistry reactions available (see Figure 1).
Figure 1: Strain-promoted Azide-Alkyne Cycloaddition (SPAAC) is a well known click-chemistry reaction. The reaction features two molecules, one functionalized with an azide group (A) and one functionalized with a cyclooctyne group (B). SPAAC is very efficient and selective, no further reagents have to be added to the reaction method and the reaction is completely bio-orthogonal. This image was adapted from last year’s iGEM team’s wiki.
As a proof of concept, iGEM TU Eindhoven 2014 employed two membrane proteins as anchors, to which they anchored DNA. These membrane proteins were CPX and INPNC. CPX in particular showed promising results and is still being explored today.
CPX & OmpX
CPX is a membrane display protein developed by Rice et al. as an alternative to traditional display methodologies, such as yeast display and phage display [2]. The protein itself is derived from the naturally occuring Outer membrane protein X (OmpX) through circular permutation (see Figure 2). Circular permutation was a necessary step to obtain the termini on the exterior of the bacterial cells.
Figure 2: CPX (right) is a membrane protein derived from OmpX (left) through circular permutation. OmpX and CPX feature signal sequences (yellow) which ensure that the membrane protein is localized to E.coli’s outer membrane. These signal sequences are cleaved from the peptides after localization. In CPX, an Amber stop codon was introduced to introduce a non-natural azide-functionalized amino acid.
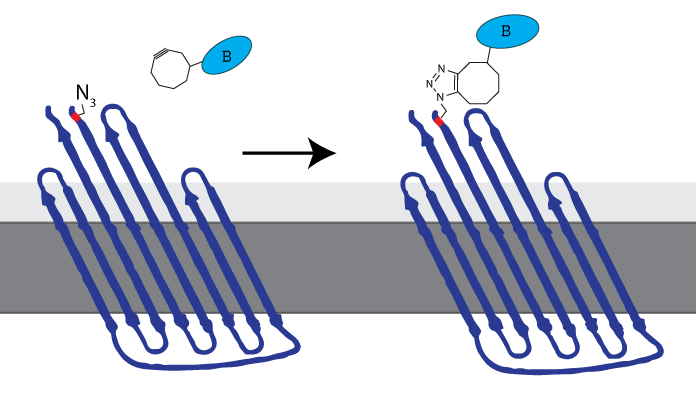
iGEM Eindhoven 2014 inserted an Amber stop codon (TAG) at the N-terminus to enable expression of an azide-functionalized amino acid. This amino-acid could be used to attach DBCO-functionalized molecules to the membrane protein, allowing synthetic biologists to attach virtually any molecule to the cell membrane (see Figure 3). The now azide-functionalized CPX-protein was dubbed Clicker Outer Membrane Protein x (COMPx) by iGEM TU Eindhoven 2014.
Figure 3: The azide-functionalized amino acid enables clicking any DBCO-functionalized molecule to be attached to the cell membrane through SPAAC-click chemistry. iGEM TU Eindhoven 2014 called this azide-functionalized CPX COMPx.
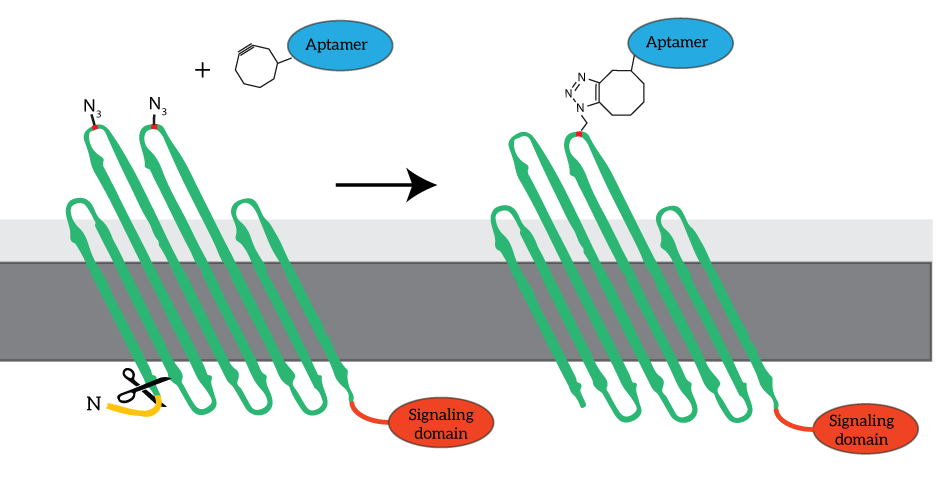
To enable signaling over the outer membrane, our system requires intracellular signaling domains. Unfortunately, the COMPx developed and characterized by last year’s iGEM team does not allow for fusing protein domains intracellularly. To obtain fusion domains with additional intracellular domains, we devised to revert to go back to the basis and use OmpX rather than CPX as the platform for our clickable outer membrane proteins. The azide-functionalized amino acid will be introduced in the loops of OmpX, to which the aptamers can be attached (see Figure 4).
Figure 4: For our signaling proteins, we revert to the basis: OmpX. We functionalize OmpX by introducing the Amber stop codon into its loops (red). The signaling domains are fused to OmpX C-terminally, since the N-terminus is occupied by the signaling sequence (yellow). The signaling sequence is cleaved from OmpX after localization.
The Loops
The Amber stop codon will be incorporated in the protruding loops of OmpX. We have chosen to introduce the mutations into the loops since they are easily accessible and are not a part of the beta-barrel of OmpX [2]. Hence, we believe that mutations in the loops will not disturb the secondary structure of OmpX (see Figure 5B).
Figure 5: A) The OmpX protein structure has been elucidated through NMR and X-ray crystallography, B) the square residues are important for the secondary structure of OmpX. To keep the structure intact, we introduce an amber stop codon in one of the protruding loops. Figure 5B is adapted from [3].
The to be substituted residues in the loops were chosen in such a way that the clicked aptamer would protrude from OmpX. Moreover, residues were selected such that the signaling proteins structure closely resembles OmpX natural’s structure. The serine residue in loop 2 was chosen since it was the only available residue with side-groups pointing outward from OmpX. The tyrosine residue in loop 3 was chosen both because tyrosine closely resembles the non-natural azide-functionalized amino acid (see Figure 6) and because the tyrosine points outward from OmpX.
Figure 6: The non-natural azide-functionalized amino acid closely resembles tyrosine
References
[1] R. Koebnik, K. P. Locher, and P. Van Gelder, “Structure and function of bacterial outer membrane proteins: barrels in a nutshell.,” Mol. Microbiol., vol. 37, no. 2, pp. 239–53, Jul. 2000.
[2] J. J. Rice, A. Schohn, P. H. Bessette, K. T. Boulware, and P. S. Daugherty, “Bacterial display using circularly permuted outer membrane protein OmpX yields high affinity peptide ligands.,” Protein Sci., vol. 15, no. 4, pp. 825–36, Apr. 2006.
[3] J. Vogt and G. E. Schulz, “The structure of the outer membrane protein OmpX from Escherichia coli reveals possible mechanisms of virulence.,” Structure, vol. 7, no. 10, pp. 1301–9, Oct. 1999.