Difference between revisions of "Part:BBa K1824559"
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1824559 short</partinfo> | <partinfo>BBa_K1824559 short</partinfo> | ||
| − | This is a specially designed | + | This is a specially designed FourU RNA thermometer that with a unique spacer at the front, which would make it compatible with promoter <partinfo>BBa_J23119</partinfo>. |
| − | + | ||
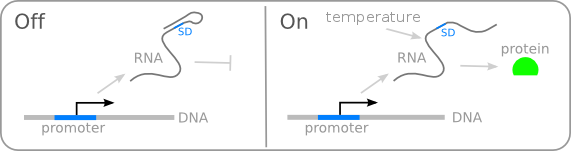
| − | Different promoters have their own transcription start sites and, in most cases, + 1 sites are embedded in promoter sequence. Hence, it is normal that transcribed RNA usually carry part of promoter sequence. However, for regulatory parts like RNA thermometer, truncation or alteration of the RNA sequence could be destructive. Hence, special designed RNA spacer between transcribed part of promoters and RNA thermometers are important for maintaining the secondary structure of RNA thermometer. | + | FourU RNA thermometer have the hairpin structure that harbors the Shine-Dalgarno sequence (SD sequence) and, in this way, make it inaccessible to the 30S unit of the bacterial ribosome, resulting in translational inactivation (Figure 2). The melting temperature of this RNA thermometer is 37 Celsius degree. Once reaching the melting temperature, hairpin structure would vanish and as a result, exposing the SD sequence to trigger the translation process. |
| − | For | + | |
| − | The possible secondary structure of | + | Different promoters have their own transcription start sites and, in most cases, + 1 sites are embedded in promoter sequence. Hence, it is normal that transcribed RNA usually carry part of promoter sequence. However, for regulatory parts like RNA thermometer, truncation or alteration of the RNA sequence could be destructive. Hence, special designed RNA spacer between transcribed part of promoters and RNA thermometers are important for maintaining the secondary structure of RNA thermometer. |
| + | |||
| + | For <partinfo>J23119</partinfo>, transcription starts at TAATGCTAGC'''A''' (transcription start site indicated in bold). Based on this, BBa_K1824559 was specially designed with a spacer that had less probability to interact with the functional structure of RNA thermometer. | ||
| + | |||
| + | The possible secondary structure of FourU was simulated by RNAstructure (Fig.1). For testing results of J23119-FourU, See <partinfo>BBa_K1824001</partinfo>. | ||
| + | |||
| + | [[Image:XJTLU-FourU.jpg|450px]] | ||
| + | [[Image:RNA_thermometer.png|450px]] | ||
| + | <br>'''Left picture: XJTLU-CHINA (2015) Figure 1:''' Possible secondary structure of RNAT FourU. | ||
| + | <br>''' Right picture: TuDelft (2008) Figure 2:''' Responsiveness of mRNA structures to environmental cues. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
Latest revision as of 15:41, 5 September 2015
RNA Thermometer FourU (Specially designed for J23119)
This is a specially designed FourU RNA thermometer that with a unique spacer at the front, which would make it compatible with promoter BBa_J23119.
FourU RNA thermometer have the hairpin structure that harbors the Shine-Dalgarno sequence (SD sequence) and, in this way, make it inaccessible to the 30S unit of the bacterial ribosome, resulting in translational inactivation (Figure 2). The melting temperature of this RNA thermometer is 37 Celsius degree. Once reaching the melting temperature, hairpin structure would vanish and as a result, exposing the SD sequence to trigger the translation process.
Different promoters have their own transcription start sites and, in most cases, + 1 sites are embedded in promoter sequence. Hence, it is normal that transcribed RNA usually carry part of promoter sequence. However, for regulatory parts like RNA thermometer, truncation or alteration of the RNA sequence could be destructive. Hence, special designed RNA spacer between transcribed part of promoters and RNA thermometers are important for maintaining the secondary structure of RNA thermometer.
For BBa_J23119, transcription starts at TAATGCTAGCA (transcription start site indicated in bold). Based on this, BBa_K1824559 was specially designed with a spacer that had less probability to interact with the functional structure of RNA thermometer.
The possible secondary structure of FourU was simulated by RNAstructure (Fig.1). For testing results of J23119-FourU, See BBa_K1824001.
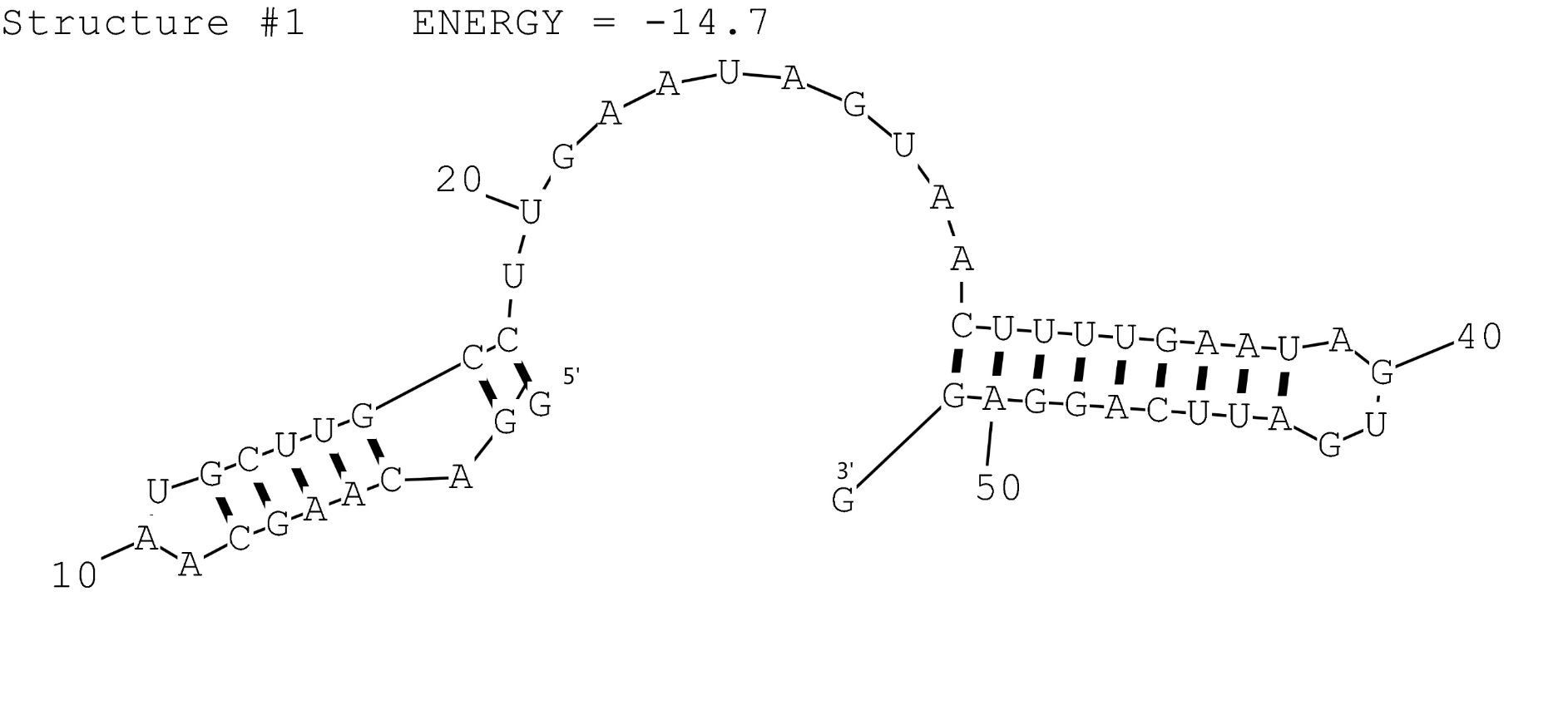
Left picture: XJTLU-CHINA (2015) Figure 1: Possible secondary structure of RNAT FourU.
Right picture: TuDelft (2008) Figure 2: Responsiveness of mRNA structures to environmental cues.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
