Difference between revisions of "Assembly:Standard assembly"
Jamesbrown (Talk | contribs) |
|||
| Line 4: | Line 4: | ||
|width=300px| | |width=300px| | ||
BioBricks have been designed to be assembled using normal cloning techniques. | BioBricks have been designed to be assembled using normal cloning techniques. | ||
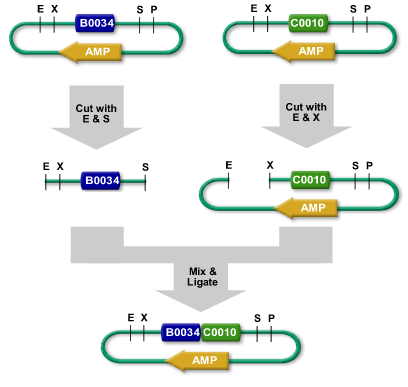
| − | Two BioBrick parts, for example one blue and one green, can be assembled into a blue-green system by a process called '''BioBrick Standard Assembly''', illustrated | + | Two BioBrick parts, for example, one blue and one green, can be assembled into a blue-green system by a process called '''BioBrick Standard Assembly''', illustrated at the left. |
| − | The BioBrick is flanked by [[Assembly:Restriction enzymes|restriction sites]], | + | The BioBrick is flanked by [[Assembly:Restriction enzymes|restriction sites]], comprised of the [[Help:BioBrick Prefix and Suffix|BioBrick Prefix and Suffix]], between the EcoRI and XbaI cutting sites on the left and SpeI and PstI on the right. For the assembly shown, the blue part is cut out of its plasmid with the enzymes EcoRI and SpeI. The resulting fragment is called the insert because it will be inserted into the [[Help:Plasmids|plasmid]] containing the other part. |
In a separate reaction, a hole is cut in the [[Help:Plasmids|plasmid]] containing the green part using EcoRI and XbaI. Using [[Help:At_the_Bench#Using_Your_BioBricks_at_the_Bench|gel electrophoresis]], the insert for the blue part and the cut plasmid containing the green part are purified and the unwanted fragments discarded. The purified insert and cut plasmid are mixed under the right conditions to allow the E sticky ends to come together and the S sticky ends to come together with the X sticky ends. Once this happens, the DNA backbone is re-ligated and the resulting blue-green part can be transformed into E.coli cells. These cells may be grown to produce as much of blue.green part as desired. | In a separate reaction, a hole is cut in the [[Help:Plasmids|plasmid]] containing the green part using EcoRI and XbaI. Using [[Help:At_the_Bench#Using_Your_BioBricks_at_the_Bench|gel electrophoresis]], the insert for the blue part and the cut plasmid containing the green part are purified and the unwanted fragments discarded. The purified insert and cut plasmid are mixed under the right conditions to allow the E sticky ends to come together and the S sticky ends to come together with the X sticky ends. Once this happens, the DNA backbone is re-ligated and the resulting blue-green part can be transformed into E.coli cells. These cells may be grown to produce as much of blue.green part as desired. | ||
|- | |- | ||
|colspan=2| | |colspan=2| | ||
| − | Each cycle of BioBrick assembly joins two parts together and in theory should only take a few | + | Each cycle of BioBrick assembly joins two parts together and in theory should only take a few hours to perform. However, less experienced assemblers may find the procedure takes a couple of days to a week. Sometimes, the assembly process fails to produce the right assembly or even to produce anything. Such results may come about because the product is not stable in ''E.coli'' and gets rejected or mutated by the cells. |
An alternative to BioBrick Standard Assembly is [http://openwetware.org/wiki/Synthetic_Biology:BioBricks/3A_assembly Three Antibiotic (3A) Assembly]. | An alternative to BioBrick Standard Assembly is [http://openwetware.org/wiki/Synthetic_Biology:BioBricks/3A_assembly Three Antibiotic (3A) Assembly]. | ||
|} | |} | ||
Revision as of 16:04, 28 November 2006
|
BioBricks have been designed to be assembled using normal cloning techniques. Two BioBrick parts, for example, one blue and one green, can be assembled into a blue-green system by a process called BioBrick Standard Assembly, illustrated at the left. The BioBrick is flanked by restriction sites, comprised of the BioBrick Prefix and Suffix, between the EcoRI and XbaI cutting sites on the left and SpeI and PstI on the right. For the assembly shown, the blue part is cut out of its plasmid with the enzymes EcoRI and SpeI. The resulting fragment is called the insert because it will be inserted into the plasmid containing the other part. In a separate reaction, a hole is cut in the plasmid containing the green part using EcoRI and XbaI. Using gel electrophoresis, the insert for the blue part and the cut plasmid containing the green part are purified and the unwanted fragments discarded. The purified insert and cut plasmid are mixed under the right conditions to allow the E sticky ends to come together and the S sticky ends to come together with the X sticky ends. Once this happens, the DNA backbone is re-ligated and the resulting blue-green part can be transformed into E.coli cells. These cells may be grown to produce as much of blue.green part as desired. | |
|
Each cycle of BioBrick assembly joins two parts together and in theory should only take a few hours to perform. However, less experienced assemblers may find the procedure takes a couple of days to a week. Sometimes, the assembly process fails to produce the right assembly or even to produce anything. Such results may come about because the product is not stable in E.coli and gets rejected or mutated by the cells. An alternative to BioBrick Standard Assembly is [http://openwetware.org/wiki/Synthetic_Biology:BioBricks/3A_assembly Three Antibiotic (3A) Assembly]. | |