Difference between revisions of "Part:BBa K1355002"
Lunalacerda (Talk | contribs) |
Lunalacerda (Talk | contribs) (→Experiments and Results) |
||
| Line 20: | Line 20: | ||
The experiment to quantify GFP expression induced by Hg was made according to the protocol “Quantification of Green Fluorescent Protein (GFP) induced by different concentrations of mercury in Escherichia coli DH5α”. | The experiment to quantify GFP expression induced by Hg was made according to the protocol “Quantification of Green Fluorescent Protein (GFP) induced by different concentrations of mercury in Escherichia coli DH5α”. | ||
| − | DH5α transformed with BBa_K1355002 was inoculated in LM (LB with low concentration of NaCl) liquid medium with chloramphenicol and grew until the Optical Density was 0.4 to 0.6abs (measured on spectrophotometer at 600 nm wavelength). After cell growth, an aliquot of 500μl in 5 eppendorf tubes (2ml) was taken and then added mercury chloride in order to achieve the concentrations of: 0.01 µg/ml, 0.02 µg/ml, 0.1 µg/ml, 0. | + | DH5α transformed with BBa_K1355002 was inoculated in LM (LB with low concentration of NaCl) liquid medium with chloramphenicol and grew until the Optical Density was 0.4 to 0.6abs (measured on spectrophotometer at 600 nm wavelength). After cell growth, an aliquot of 500μl in 5 eppendorf tubes (2ml) was taken and then added mercury chloride in order to achieve the concentrations of: 0 µg/ml, 0.01 µg/ml, 0.02 µg/ml, 0.1 µg/ml, 0.2µg/ml and 1 µg/ml. The samples were incubated at 37°C on shaker. We collected each eppendorf tube at time 1 (01:30 hours of incubation), time 2 (03:00 hours of incubation) and time 3 (04:30 hours of incubation). Every sample was centrifuged at 12000g for 3 minutes and the pellet washed with TN Buffer (Nacl 0.15M + Tris HCl 10mM) and then re-suspended with 500μl of the same buffer. The same process was made to the bacterium without construction as a control to GFP expression/intensity. GFP expression was measured using the Hidex Chameleon spectrofluorimeter with excitation filter 340 nm and emission filter 500 nm wavelength. The Optical Density was measured simultaneously. All samples were analyzed in triplicate. |
The graph represented on Figure 1 shows the Optical Density of transformed DH5α with BBa_K1355002 in different Hg concentrations in function of time: | The graph represented on Figure 1 shows the Optical Density of transformed DH5α with BBa_K1355002 in different Hg concentrations in function of time: | ||
| − | [[File:bs1.png]] | + | [[File:bs1.png| 600px]] |
| − | '''Figure 1.''' Optical Density measured in the four given times, at mercury chloride concentrations of 0 µg/ml, 0.01 µg/ml, 0.02 µg/ml, 0.1 µg/ml, 0. | + | '''Figure 1.''' Optical Density measured in the four given times, at mercury chloride concentrations of 0 µg/ml, 0.01 µg/ml, 0.02 µg/ml, 0.1 µg/ml, 0.2µg/ml and 1 µg/ml. |
| − | + | As we can observe, after four hours of induction by mercury, GFP expression increases according to the concentration. Mercury Bacter biosensor was efficient even in concentrations as low as 10 ppb, and got still higher emissions in solutions of 1000 ppb. This versatilty makes it a good indicator of water polluted with mercury. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
===References=== | ===References=== | ||
Revision as of 16:04, 1 November 2014
Mercury ions detector device
We designed a biobrick device to express Green Fluorescent Protein in mercury’s occurrence. The Mercury ions’ detector device biobrick (BBa_K1355002) is composed by mer bidirectional promoter (BBa_K1355001) attached to the GFP translational unit (BBa_E0840) available on iGEM database since 2004 . It has dual function: A) In reverse: MerR protein regulator transcription; and B) in forward: transcription of MerP - MerT - GFP proteins, as represented below:
In absence of mercury, MerR forms a MerR-promoter-operator complex, preventing RNA polymerase to recognize the promoter, consequently, messengers RNA for MerPT and GFP will not be transcript. In presence of Hg2+, MerR protein binds to this element and dissociates from the promoter-operator complex, allowing MerPT and GFP expression, as represented below:
When MerT, MerP and the Green Fluorescent Protein (GFP) protein are expressed, the biodetection will start! MerP and MerT proteins are responsible for the transport of mercury from the periplasm to cytoplasm and GFP will act as a reporter alerting the presence of mercury in the middle and higher is mercury’s concentration, higher will be GFP expression.
Figure 1: Mercury Bacter Hg biodetector (DH5-alpha transformed with BBa_K1355002)
Experiments and Results
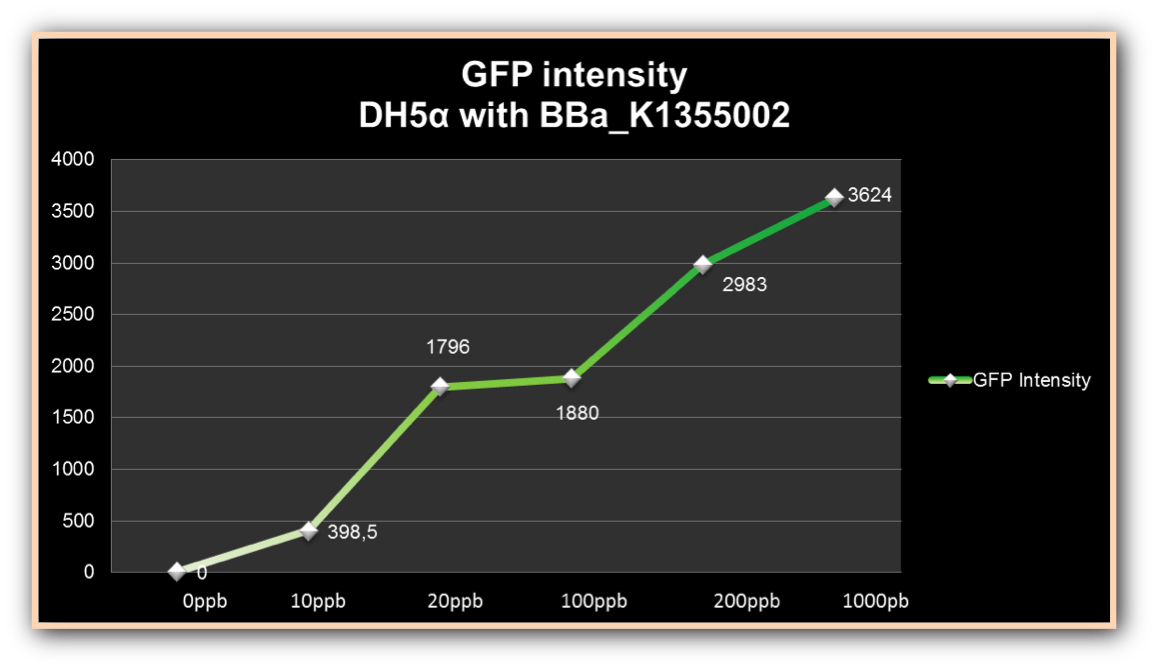
The experiment to quantify GFP expression induced by Hg was made according to the protocol “Quantification of Green Fluorescent Protein (GFP) induced by different concentrations of mercury in Escherichia coli DH5α”. DH5α transformed with BBa_K1355002 was inoculated in LM (LB with low concentration of NaCl) liquid medium with chloramphenicol and grew until the Optical Density was 0.4 to 0.6abs (measured on spectrophotometer at 600 nm wavelength). After cell growth, an aliquot of 500μl in 5 eppendorf tubes (2ml) was taken and then added mercury chloride in order to achieve the concentrations of: 0 µg/ml, 0.01 µg/ml, 0.02 µg/ml, 0.1 µg/ml, 0.2µg/ml and 1 µg/ml. The samples were incubated at 37°C on shaker. We collected each eppendorf tube at time 1 (01:30 hours of incubation), time 2 (03:00 hours of incubation) and time 3 (04:30 hours of incubation). Every sample was centrifuged at 12000g for 3 minutes and the pellet washed with TN Buffer (Nacl 0.15M + Tris HCl 10mM) and then re-suspended with 500μl of the same buffer. The same process was made to the bacterium without construction as a control to GFP expression/intensity. GFP expression was measured using the Hidex Chameleon spectrofluorimeter with excitation filter 340 nm and emission filter 500 nm wavelength. The Optical Density was measured simultaneously. All samples were analyzed in triplicate.
The graph represented on Figure 1 shows the Optical Density of transformed DH5α with BBa_K1355002 in different Hg concentrations in function of time:
Figure 1. Optical Density measured in the four given times, at mercury chloride concentrations of 0 µg/ml, 0.01 µg/ml, 0.02 µg/ml, 0.1 µg/ml, 0.2µg/ml and 1 µg/ml.
As we can observe, after four hours of induction by mercury, GFP expression increases according to the concentration. Mercury Bacter biosensor was efficient even in concentrations as low as 10 ppb, and got still higher emissions in solutions of 1000 ppb. This versatilty makes it a good indicator of water polluted with mercury.
References
Farias, L. A., Fávaro, D. I., Pessoa, A., Aguiar, J. P., & Yuyama, L. K. (2012). Mercury and methylmercury concentration assessment in children's hair from Manaus, Amazonas state, Brazil. Acta Amazonica, 42(2), 279-286.
Fillion, M., Philibert, A., Mertens, F., Lemire, M., Passos, C. J. S., Frenette, B., ... & Mergler, D. (2011). Neurotoxic sequelae of mercury exposure: an intervention and follow-up study in the Brazilian Amazon. Ecohealth, 8(2), 210-222.
Grotto, D., Valentini, J., Fillion, M., Passos, C. J. S., Garcia, S. C., Mergler, D., & Barbosa Jr, F. (2010). Mercury exposure and oxidative stress in communities of the Brazilian Amazon. Science of the Total Environment, 408(4), 806-811.
Hakkila, K., Maksimow, M., Karp, M., & Virta, M. (2002). Reporter Genes lucFF, luxCDABE, gfp, and dsred Have Different Characteristics in Whole-Cell Bacterial Sensors. Analytical biochemistry, 301(2), 235-242.
Kuncova, G., Pazlarova, J., Hlavata, A., Ripp, S., & Sayler, G. S. (2011). Bioluminescent bioreporters Pseudomonas putida TVA8 as a detector of water pollution. Operational conditions and selectivity of free cells sensor. Ecological Indicators, 11(3), 882-887.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 988
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 586
Illegal NgoMIV site found at 1160 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1862
Illegal SapI site found at 579