Difference between revisions of "Part:BBa K1321306"
| Line 10: | Line 10: | ||
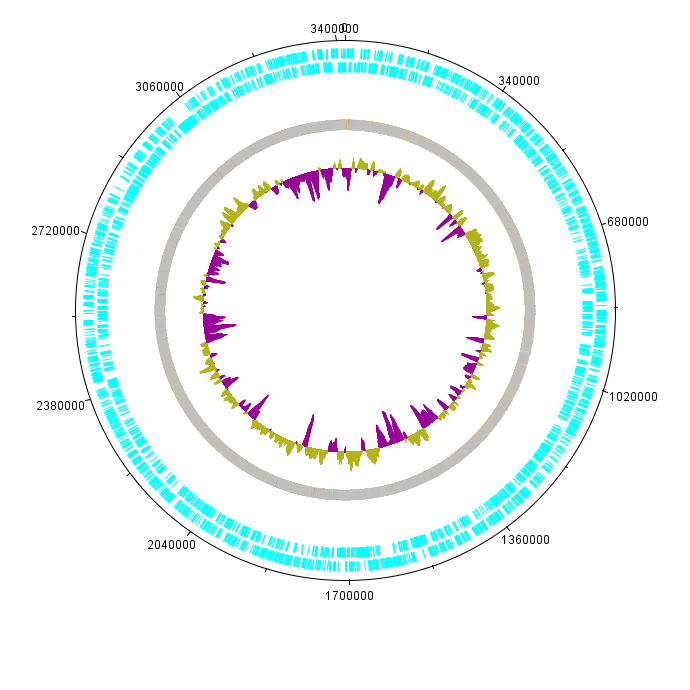
G.xylinus igem genome was sequenced as a part of Imperial iGEM 2014 project. The genome is approximately 3.4Mbp in size, with a GC content of 63.52% (see Figure 1). Autoannotation using Prokka (Seemann 2014) identified in total 3052 putative coding sequences, 59 tRNA sequences and 24 rRNA and other RNA sequences. | G.xylinus igem genome was sequenced as a part of Imperial iGEM 2014 project. The genome is approximately 3.4Mbp in size, with a GC content of 63.52% (see Figure 1). Autoannotation using Prokka (Seemann 2014) identified in total 3052 putative coding sequences, 59 tRNA sequences and 24 rRNA and other RNA sequences. | ||
| − | [[File:IC14 G.xylinus igem genome view.png|400px|thumb|left|'''Figure 1.''' Overview of the genome of G.xylinus igem. The genome is approximately | + | [[File:IC14 G.xylinus igem genome view.png|400px|thumb|left|'''Figure 1.''' Overview of the genome of G.xylinus igem. The genome is approximately 3.4Mbp in size, with a GC content of around 63% (internal circle) and approximately 3000 coding sequences (external circle; blue bands denote single CDSs).]] |
Revision as of 14:48, 22 October 2014
Komagataeibacter rhaeticus iGEM
Gluconacetobacter xylinus is a obligate aerobe Gram negative cellulose-producing bacterial species. Imperial iGEM 2014 team isolated a strain of G.xylinus (named G. xylinus igem in the honour of the competition) from the Kombucha tea seed culture (Kombucha tea seed culture is a mixture of yeast, G.xylinus and other organisms, and is used to make the increasingly popular Kombucha tea). G. xylinus igem produces cellulose readily on different carbon feedstocks. Although G. xylinus igem does not produce as high amounts of cellulose as G. xylinus ATCC 53582 (see cellulose productivity on different feedstocks), it is more amenable to genetic engineering and testing of genetic constructs. BBa_K1321306 is a member of the G.xylinus genetic engineering toolkit (parts BBa_K1321295 - BBa_K1321332).
G.xylinus igem genome sequence
G.xylinus igem genome was sequenced as a part of Imperial iGEM 2014 project. The genome is approximately 3.4Mbp in size, with a GC content of 63.52% (see Figure 1). Autoannotation using Prokka (Seemann 2014) identified in total 3052 putative coding sequences, 59 tRNA sequences and 24 rRNA and other RNA sequences.
Culturing G. xylinus igem
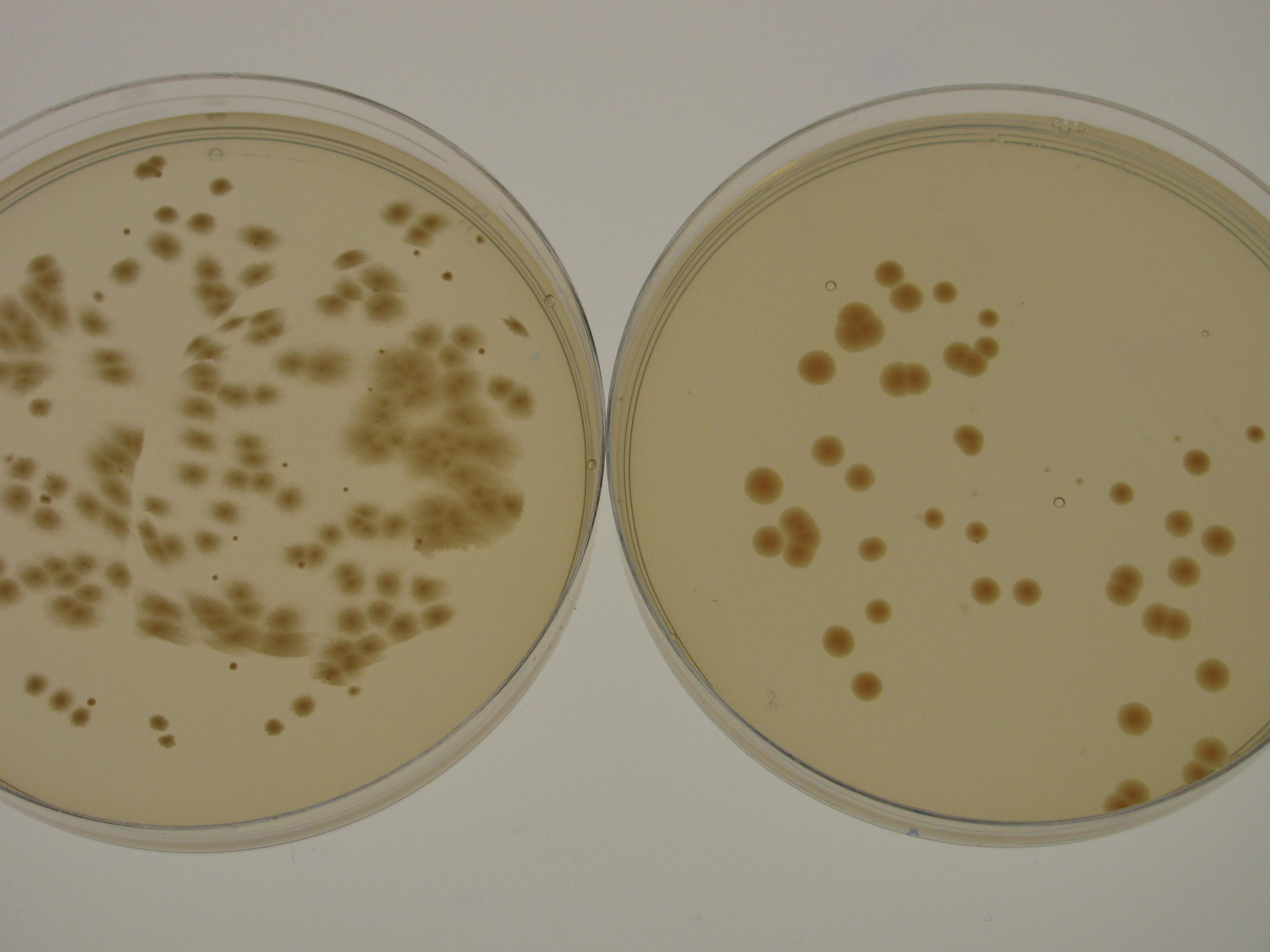
G.xylinus igem growth rate varies depending on the culturing conditions. Growth rate is highest when G.xylinus is cultured at 30degC, 180rpm shaking in a well-aerated setting. However, in shaking conditions, the prevalence of cellulose non-producing mutants (cel- mutants) increases, which causes a decrease in the total yield of produced cellulose. For this reason, when high cellulose production is required, G.xylinus is often grown in static conditions, at 30degC, in a well-aerated setting. In shaking cultures, igem strain reaches stationary phase in 48-72 hours, whereas in static cultures, depending on the volume of the medium, it can take up to 15 days. When grown on HS-agar plates at 30degC, cellulose-producing colonies become visible within 48-72 hours, cel- mutant colonies between 24 and 48 hours. Cellulose producing colonies can be distinguished from cel- mutants by colony morphology: cel+ have a rough colony morphology, whereas cel- mutants have a smooth colony morphology (see Figure 1).
The most commonly used medium for G.xylinum culturing is Hestrin-Schramm (HS) medium. HS medium contains the following components (for a detailed overview for G.xylinus culturing, see G.xylinus culturing on [http://2014.igem.org/Team:Imperial/Protocols Imperial 2014 wiki]:
2% (w/v) glucose
0.5% (w/v) yeast extract
0.5% (w/v) peptone
0.27% (w/v) Na2HPO4
0.15% (w/v) citric acid.
Cellulose productivity of G.xylinus igem
G.xylinus igem cellulose productivity depends strongly on the carbon feedstocks used. In HS-glucose media grown for 7 days at 30degC standing using 50ml HS medium in 250ml conical flasks (sealed with foam buns), cellulose productivity is approximately one fifth of that of ATCC 53582 strain (see Figure 1 for comparison of ATCC53582 and igem strains and experimental details). However, igem growth is not optimal in HS-glucose, because in HS-glycerol media and HS-sucrose media the cellulose productivity is increased. Furthermore, in HS-sucrose medium, cellulose productivity is higher than that of ATCC 53582, which is of industrial significance due to the high availability of sucrose (see Feedstocks for G.xylinus igem).
Feedstocks for G.xylinus igem
G.xylinus is commonly grown on HS-glucose. However, it can readily use other carbon feedstocks, including sucrose and glycerol. igem strain's cellulose productivity is highest when grown on glycerol, and higher than ATCC 53582 when grown on sucrose (see Figure 2). Although glycerol seems to result in highest cellulose production, the effects of routine culturing of ATCC 53582 on HS-glycerol media are unknown.
Natural antibiotics resistance of G.xylinus igem
Using optimal antibiotics concentrations with G.xylinus igem is critical for selection of transformed cells while not overly inhibiting cell growth. G.xylinus igem is naturally resistant to kanamycin, ampicillin and chloramphenicol concentrations normally used for E.coli (50µg/ml, 100µg/ml and 35µg/ml respectively; see Figure 4 for an experiment of igem natural antibiotic concentrations, experimental details, and comparison of ATCC and igem strains). The frequency of appearance of antibiotic-resistant colonies is dependent on the number of cells used for plating - with higher number of plated cells, colonies appear even at higher antibiotics concentrations. Imperial iGEM 2014 team found that igem strain forms antibiotic-resistant colonies up to 6x antibiotic concentrations (6x of those commonly used for E.coli), thus the recommended amounts of antibiotics for HS-plates after transformations are 350µg/ml kanamycin, 700µg/ml ampicillin and 245µg/ml chloramphenicol.
NOTE: Because the registry's standard plasmid backbone pSB1C3 is not capable of replication in Gluconacetobacter species, the G.xylinus genetic engineering toolkit is housed mainly in pSEVA331-Bb. pSEVA331-Bb is a non-standard backbone, which therefore can't be quality controlled by and maintained in the Registry. However, in order to make the G.xylinus toolkit available for the synthetic biology community, Imperial iGEM 2014 team has made it freely available upon request, with quality control provided (see Experience). To request, please contact Imperial iGEM 2014 team.
References:
Seemann, T. (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics (Oxford, England). [Online] 30 (14), 2068–2069. Available from: doi:10.1093/bioinformatics/btu153 [Accessed: 10 July 2014].
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]