Difference between revisions of "Part:BBa K1372001"
| Line 2: | Line 2: | ||
<partinfo>BBa_K1372001 short</partinfo> | <partinfo>BBa_K1372001 short</partinfo> | ||
| − | This | + | This Biobrick is the result of the assembly of two BB: |
- [https://parts.igem.org/Part:BBa_K1372000 BBa_K1372000]: | - [https://parts.igem.org/Part:BBa_K1372000 BBa_K1372000]: | ||
| Line 14: | Line 14: | ||
[[File:Paris Saclay BB2001.png|center]] | [[File:Paris Saclay BB2001.png|center]] | ||
| − | This | + | This Bioibrick could thus be used as control system of the production of tRNA-SUPD depending on the concentration of a substrate (salicylate) gradually consumed by the organisms in the growth-medium. Consequentially, the production of tRNA-SupD will gradually peter out. |
An example of use of this control system is described there: [http://2014.igem.org/Team:Paris_Saclay/Project/Salicylate_Inducible_System Salicylate Inducible System] | An example of use of this control system is described there: [http://2014.igem.org/Team:Paris_Saclay/Project/Salicylate_Inducible_System Salicylate Inducible System] | ||
Revision as of 02:08, 18 October 2014
Salicylate Inducible Suppressing System
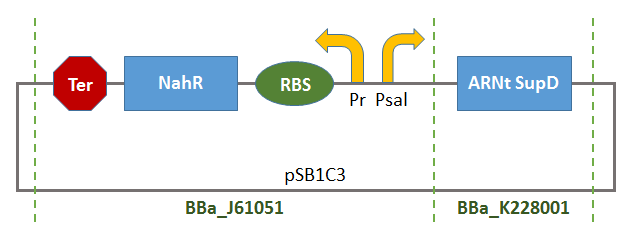
This Biobrick is the result of the assembly of two BB:
- BBa_K1372000:
- BBa_B0015: Double terminator consisting of BBa_B0010 and BBa_B0012.
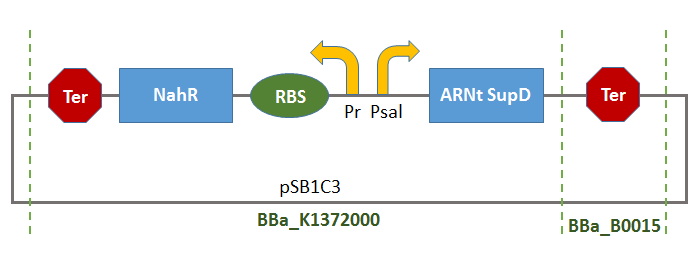
Thus, there will be a terminator of the transcription of tRNA-SUPD gene (missing in BBa_K1372000).
This Bioibrick could thus be used as control system of the production of tRNA-SUPD depending on the concentration of a substrate (salicylate) gradually consumed by the organisms in the growth-medium. Consequentially, the production of tRNA-SupD will gradually peter out.
An example of use of this control system is described there: [http://2014.igem.org/Team:Paris_Saclay/Project/Salicylate_Inducible_System Salicylate Inducible System]
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 786
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 77
Illegal NgoMIV site found at 618
Illegal AgeI site found at 1309 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1312
Made by [http://2014.igem.org/Team:Paris_Saclay iGEM Paris Saclay]