Difference between revisions of "Part:BBa K1554002"
(→Usage and Biology) |
|||
| (7 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1554002 short</partinfo> | <partinfo>BBa_K1554002 short</partinfo> | ||
| − | HarFAR-3 fatty acid reductase from ''Helicoverpa armigera'' with an ER retention signal (KKYR) attached to the C-terminal end | + | HarFAR-3 fatty acid reductase from ''Helicoverpa armigera'' with an ER retention signal (KKYR) attached to the C-terminal end. |
===Usage and Biology=== | ===Usage and Biology=== | ||
The HarFAR protein is a fatty acid reductase coming from ''Helicoverpa armigera'' which transforms fatty acids into fatty alcohols. | The HarFAR protein is a fatty acid reductase coming from ''Helicoverpa armigera'' which transforms fatty acids into fatty alcohols. | ||
| − | + | ||
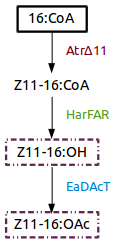
| + | [https://parts.igem.org/Part:BBa_K1554001 Part:BBa_K1554001] (AtrΔ11), [https://parts.igem.org/Part:BBa_K1554002 Part:BBa_K1554002] (HarFAR) and [https://parts.igem.org/Part:BBa_K1554003 Part:BBa_K1554003] (EaDAcT) are enzymes of a biosynthesis pathway that lead to the production of insect sexual pheromones, Z11-16:OH and Z11-16:OAc, using palmitate as substrate. | ||
| + | |||
| + | |||
| + | [[File:VUPV_pathway_parts.png]] | ||
| + | |||
| + | Figure 1. Insect sexual pheromone pathway for ''Nicotiana benthamiana''. | ||
| + | |||
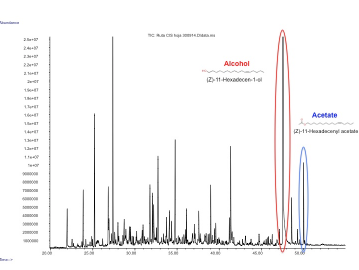
| + | In our project we made a device with these three pheromones and expressed them by transient expression it in our plant chasis, ''Nicotiana benthamiana''. In order to check if the insect sexual pheromones were present, we performed the analysis using HS-SPME coupled to GC-MS. We observed two additional peaks in the transformed plants that were not present in the control and had a similar mass spectrum and retention time as the standards, which confirmed that both molecules were the desired pheromones, (Z)-11-hexadecen-1-ol and (Z)-11-hexadecenyl acetate. | ||
| + | |||
| + | [[File:UPV_rutas-biosintesis_feromonas.png]] | ||
| + | |||
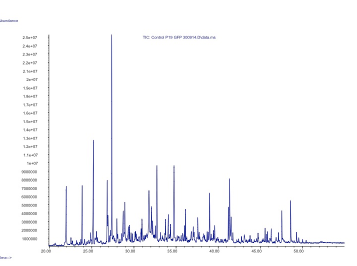
| + | Figure 2. GC-MS analysis of the volatile organic compounds from a negative control of ''Nicotiana benthamiana''. | ||
| + | |||
| + | [[File:VUPV_pheromone.png]] | ||
| + | |||
| + | Figure 3. GC-MS analysis of the volatile organic compounds from a genetically engineered ''Nicotiana benthamiana'' to produce insect pheromones. | ||
| + | |||
| + | |||
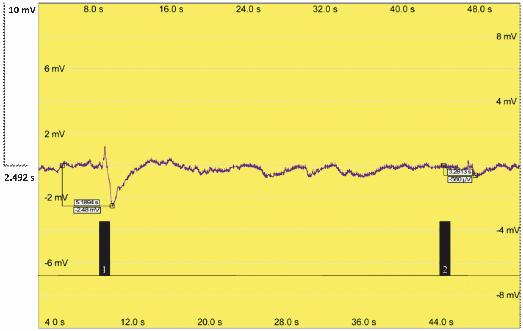
| + | Additionally, our team performed an electroantennography (EAG) to test the moth response to pheromones. We connected one antenna from a male moth, ''Sesamia nonagrioides'', with the two electrodes. Then, an air current with a leaf extract containing our pheromones was applied. As the extract was applied, the antenna transmitted an electrical impulse, meaning that there was response to our insect pheromones produced in plant. The volatiles in our plants induced detectable electric pulses that could indicate a pheromone response. | ||
| + | |||
| + | |||
| + | [[File:UPV_EAG.png]] | ||
| + | |||
| + | Figure 4. Electroantennography analysis of Sesamia nonagroides response to sexual pheromones produced in genetically engineered ''Nicotiana Benthamiana plants''. Signal 1: Antennal response to the Sexy Plant leaf extract. Signal 2: Antennal response to an air puff. | ||
| + | |||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
Latest revision as of 17:38, 17 October 2014
HarFAR
HarFAR-3 fatty acid reductase from Helicoverpa armigera with an ER retention signal (KKYR) attached to the C-terminal end.
Usage and Biology
The HarFAR protein is a fatty acid reductase coming from Helicoverpa armigera which transforms fatty acids into fatty alcohols.
Part:BBa_K1554001 (AtrΔ11), Part:BBa_K1554002 (HarFAR) and Part:BBa_K1554003 (EaDAcT) are enzymes of a biosynthesis pathway that lead to the production of insect sexual pheromones, Z11-16:OH and Z11-16:OAc, using palmitate as substrate.
Figure 1. Insect sexual pheromone pathway for Nicotiana benthamiana.
In our project we made a device with these three pheromones and expressed them by transient expression it in our plant chasis, Nicotiana benthamiana. In order to check if the insect sexual pheromones were present, we performed the analysis using HS-SPME coupled to GC-MS. We observed two additional peaks in the transformed plants that were not present in the control and had a similar mass spectrum and retention time as the standards, which confirmed that both molecules were the desired pheromones, (Z)-11-hexadecen-1-ol and (Z)-11-hexadecenyl acetate.
Figure 2. GC-MS analysis of the volatile organic compounds from a negative control of Nicotiana benthamiana.
Figure 3. GC-MS analysis of the volatile organic compounds from a genetically engineered Nicotiana benthamiana to produce insect pheromones.
Additionally, our team performed an electroantennography (EAG) to test the moth response to pheromones. We connected one antenna from a male moth, Sesamia nonagrioides, with the two electrodes. Then, an air current with a leaf extract containing our pheromones was applied. As the extract was applied, the antenna transmitted an electrical impulse, meaning that there was response to our insect pheromones produced in plant. The volatiles in our plants induced detectable electric pulses that could indicate a pheromone response.
Figure 4. Electroantennography analysis of Sesamia nonagroides response to sexual pheromones produced in genetically engineered Nicotiana Benthamiana plants. Signal 1: Antennal response to the Sexy Plant leaf extract. Signal 2: Antennal response to an air puff.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1104
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI site found at 929