Difference between revisions of "Part:BBa K1355000"
Lunalacerda (Talk | contribs) (→Source) |
Lunalacerda (Talk | contribs) |
||
| Line 17: | Line 17: | ||
MerA gene sequence is found in the O26-CRL plasmid from Escherichia coli O26. We also added strong RBS from protein 10 found in phage 17 and the double terminator (BBa_B0015) to ensure efficient termination of transcription and messenger RNA recognition. | MerA gene sequence is found in the O26-CRL plasmid from Escherichia coli O26. We also added strong RBS from protein 10 found in phage 17 and the double terminator (BBa_B0015) to ensure efficient termination of transcription and messenger RNA recognition. | ||
| + | ===References=== | ||
| + | Barkay, T., Miller, S. M., & Summers, A. O. (2003). Bacterial mercury resistance from atoms to ecosystems. FEMS microbiology reviews, 27(2‐3), 355-384. | ||
| + | |||
| + | BIONDO, R. Engenharia Genética de Cupriavidus metallidurans CH34 para a Biorremediação de efluentes contendo Metais Pesados. 2008. São Paulo, Brasil. | ||
| + | |||
| + | Nascimento, A. M., & Chartone-Souza, E. (2003). Operon mer: bacterial resistance to mercury and potential for bioremediation of contaminated environments. Genetics and Molecular Research, 2(1), 92-101. | ||
| + | |||
| + | DASH, H. R.; DAS, S. Bioremediation of mercury and the importance of bacterial mer genes. 2012. International Biodeterioration & Biodegradation 75: 207 – 213. | ||
| + | |||
| + | HAMLETT, N. V.; et al. Roles of the Tn21 merT, merP and merC Gene Products in Mercury Resistance and Mercury Binding. 1992. Journal of Bacteriology 174: 6377 – 6385. | ||
| + | |||
| + | PINTO, M. N. Bases Moleculares da resistência ao Mercúrio em bactérias gram-negativas da Amazônia brasileira. 2004. Pará, Brasil. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 15:35, 17 October 2014
Strong RBS + merA (mercuric ion reductase)+ terminator (BBa_ B0015)
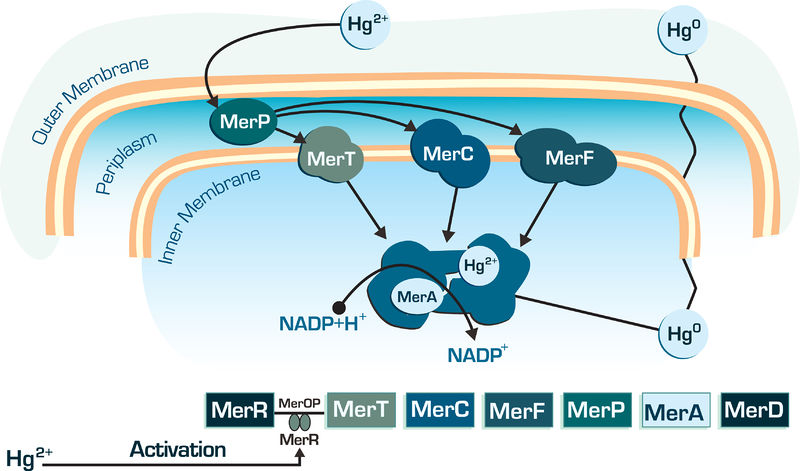
The MerA gene is one of the most important gene into bacterial mercury-resistance operon. The mercuric reductase is a cytosolic flavin disulfide oxidoreductase (homodimer ∼120 kDa) which uses NAD(P)H as a reductant. Transforms mercury Hg2+ in Hg0, which volatile, lipid-soluble Hg(0) diffuses through the cell membranes without the need for any dedicated efflux system, as shown in figure below.
Figure 01: Mer genes action in bacterial cell
Figure 02: MerA homodimer protein 3D structure
Source
MerA gene sequence is found in the O26-CRL plasmid from Escherichia coli O26. We also added strong RBS from protein 10 found in phage 17 and the double terminator (BBa_B0015) to ensure efficient termination of transcription and messenger RNA recognition.
References
Barkay, T., Miller, S. M., & Summers, A. O. (2003). Bacterial mercury resistance from atoms to ecosystems. FEMS microbiology reviews, 27(2‐3), 355-384.
BIONDO, R. Engenharia Genética de Cupriavidus metallidurans CH34 para a Biorremediação de efluentes contendo Metais Pesados. 2008. São Paulo, Brasil.
Nascimento, A. M., & Chartone-Souza, E. (2003). Operon mer: bacterial resistance to mercury and potential for bioremediation of contaminated environments. Genetics and Molecular Research, 2(1), 92-101.
DASH, H. R.; DAS, S. Bioremediation of mercury and the importance of bacterial mer genes. 2012. International Biodeterioration & Biodegradation 75: 207 – 213.
HAMLETT, N. V.; et al. Roles of the Tn21 merT, merP and merC Gene Products in Mercury Resistance and Mercury Binding. 1992. Journal of Bacteriology 174: 6377 – 6385.
PINTO, M. N. Bases Moleculares da resistência ao Mercúrio em bactérias gram-negativas da Amazônia brasileira. 2004. Pará, Brasil.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1200
Illegal NgoMIV site found at 1262 - 1000COMPATIBLE WITH RFC[1000]