Difference between revisions of "Part:BBa K1355000"
Lunalacerda (Talk | contribs) |
Mctastolfi (Talk | contribs) |
||
| Line 1: | Line 1: | ||
<partinfo>BBa_K1355000 short</partinfo> | <partinfo>BBa_K1355000 short</partinfo> | ||
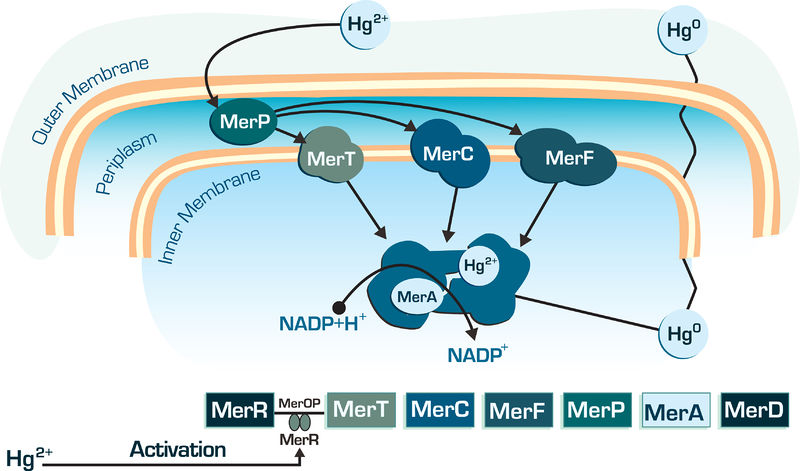
| − | The MerA gene is one of the most important gene into bacterial mercury-resistance operon. Transforms mercury Hg2+ in Hg0, which | + | The MerA gene is one of the most important gene into bacterial mercury-resistance operon. The mercuric reductase is a cytosolic flavin disulfide oxidoreductase (homodimer ∼120 kDa) which uses NAD(P)H as a reductant. Transforms mercury Hg2+ in Hg0, which volatile, lipid-soluble Hg(0) diffuses through the cell membranes without the need for any dedicated efflux system, as shown in figure below. |
| Line 7: | Line 7: | ||
Figure 01: Mer genes action in bacterial cell | Figure 01: Mer genes action in bacterial cell | ||
| − | + | ||
[[File:l2.png]] | [[File:l2.png]] | ||
| − | Figure 02: MerA protein 3D | + | Figure 02: MerA homodimer protein 3D structure |
== Source == | == Source == | ||
Revision as of 14:44, 13 October 2014
Strong RBS + merA (mercuric ion reductase)+ terminator (BBa_ B0015)
The MerA gene is one of the most important gene into bacterial mercury-resistance operon. The mercuric reductase is a cytosolic flavin disulfide oxidoreductase (homodimer ∼120 kDa) which uses NAD(P)H as a reductant. Transforms mercury Hg2+ in Hg0, which volatile, lipid-soluble Hg(0) diffuses through the cell membranes without the need for any dedicated efflux system, as shown in figure below.
Figure 01: Mer genes action in bacterial cell
Figure 02: MerA homodimer protein 3D structure
Source
MerA gene sequence is found in the O26-CRL plasmid from Escherichia coli O26. We also added strong RBS from protein 10 found in phage 17 and the double terminator (BBa_B0015) to ensure efficient termination of transcription and messenger RNA recognition.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1200
Illegal NgoMIV site found at 1262 - 1000COMPATIBLE WITH RFC[1000]