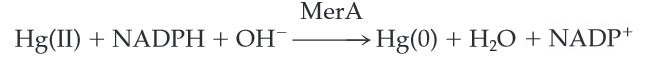Difference between revisions of "Part:BBa K1420001"
(→Characterization of merA) |
(→Overview) |
||
| Line 3: | Line 3: | ||
== Overview == | == Overview == | ||
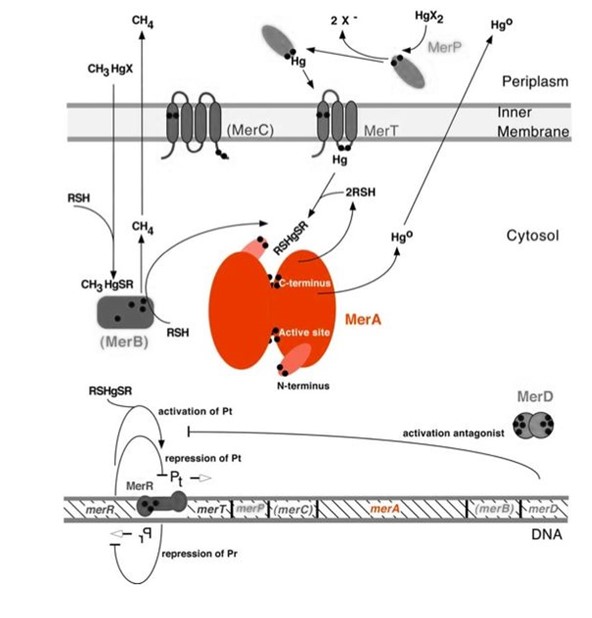
| − | <p>Mercury resistance gene ''merA'' ( | + | <p>Mercury resistance gene ''merA'' (1686 bp) encodes MerA, a mercuric ion reductase (cytosolic, flavin disulfide oxidoreductase, ~120kDa), that is essential to bacterial mercury resistance. MerA and gene ''merA'' are highlighted in orange in Figure 1. The lower part of the Figure 1 shows the arrangement of ''mer'' genes in the operon, and ''merA'' is located downstream of ''merT'' and ''merP'', the genes that encode mercury transport proteins. As shown in ''Scheme 1'', the essential mechanism of mercury resistance is carried out by MerB and MerA. MerB transforms methylmercury to less toxic, non-biomagnified ionic mercury Hg(II), and MerA reduces Hg(II) to the least toxic metallic mercury, Hg(0).</p> |
[[File:MerA.jpg|center]] | [[File:MerA.jpg|center]] | ||
Revision as of 02:58, 13 October 2014
merA, mercuric reductase from Serratia marcescens
Overview
Mercury resistance gene merA (1686 bp) encodes MerA, a mercuric ion reductase (cytosolic, flavin disulfide oxidoreductase, ~120kDa), that is essential to bacterial mercury resistance. MerA and gene merA are highlighted in orange in Figure 1. The lower part of the Figure 1 shows the arrangement of mer genes in the operon, and merA is located downstream of merT and merP, the genes that encode mercury transport proteins. As shown in Scheme 1, the essential mechanism of mercury resistance is carried out by MerB and MerA. MerB transforms methylmercury to less toxic, non-biomagnified ionic mercury Hg(II), and MerA reduces Hg(II) to the least toxic metallic mercury, Hg(0).
Figure 1. Model of mercury resistance operon. The symbol • indicates a cysteine residue. RSH indicates cytosolic thiol redox buffers such as glutathione. Figure 1 shows the interactions of MerA with mercury compounds and other gene products of mer operon. (This figure is adapted from "Bacterial mercury resistance from atoms to ecosystems". Reference: T. Barkay et al. FEMS Microbiology Reviews 27 (2003) 355-384.)
The nature evolves different mercury detoxification strategies. Here we describe MerA and MerB confer mercury resistance by coupling electrochemical reductions of thiol-avid organic mercury species to relatively inert, uncharged, monoatonic mercury. Since organic mercuric species have high affinity to tiol groups and commomly form strong but reversible bonds,cytosolic thiol redox buffers and cysteine residues of mercury binding proteins compete with thiol groups in other protein and therefore prevents cationic mercury from inhibiting cytosolic machineries. Once the organomercury is converted to ionic mercury by MerB, MerA reduces ionic mercury to volatile, elemental mercury which diffuses through cell membrane without any active transport systems. While MerA and MerB are the key mercury detoxification enzymes, they orchestrate with mercury transport proteins, MerT and MerP, to confer bacterial mercury resistance.
Molecular Function
MerA catalyzes the reduction of mercuric ion to the relative inert, volatile monoatomic mercury in a NADPH dependent reaction. Ionic mercury Hg(II) is released from protonlysis of organomercury catalyzed by MerB.
Scheme 1. MerA Catalyzed Reaction
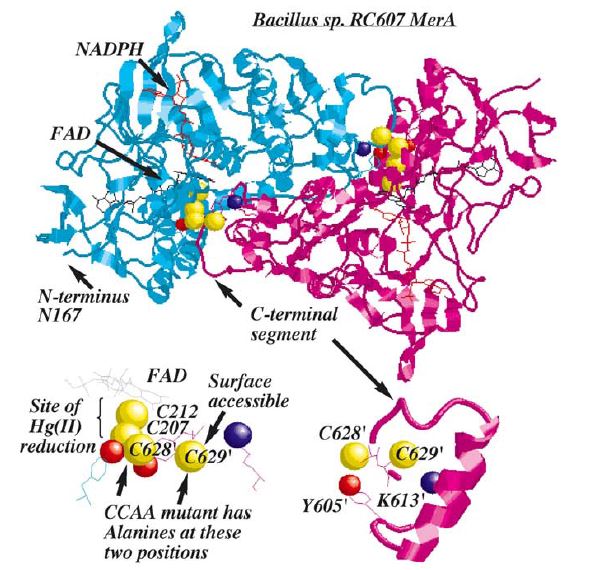
Structure and Mechanism
MerA is a 125kDA homodimer.
Characterization of merA
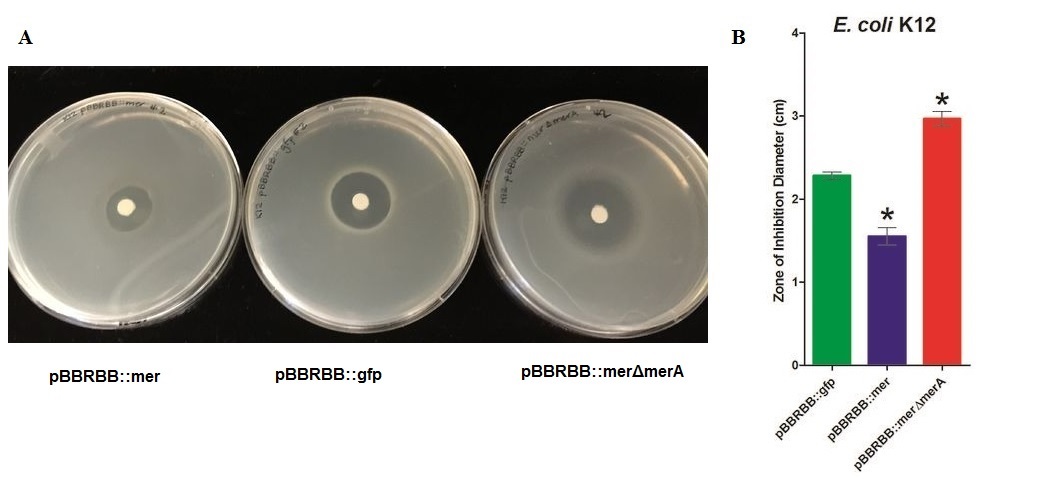
To test the effect of MerA to mercury resistance, we generated a merA deletion construct from pBBRBB::mer and characterized it by zone of inhibition test.
Zone of Inhibition Results
Figure 2. Zones of Inhibition Test For Mercury Resistance Activity. In this assay, Escherichia coli strain K12 with three different constructs expressed were spread on agar plate to compare level of mercury resistance. Each agar plate had a filter disk spotted with 10µL of 0.1M HgCl2 in the middle . (A)Agar Plates of K12 with each construct. Left, K12 conntaining pBBRBB::mer (mer operon); center,K12 containing pBBRBB::gfp (empty vector); right, K12 containing pBBRBB::merΔmerA(mer operon with merA deletion mutant). (b) Size of zones of inhibition diameter. The diameter of the Zone of Inhibition was measured in triplicate. Green corresponds to pBBRBB::gfp, blue to pBBRBB::mer, and red to pBBRBB::merΔmerA. Zone of inhibition of individual construct arranges from highest inhibition to lowest inhibition is: merΔmerA > empty vector > mer.
In the E. coli K12 with the empty vector had higher zone of inhibition than the E. coli with mer operon. In addition, vector with MerA deleted has a highest zone of inhibition among three samples, which would be expected as the bacteria would be unable to reduce Hg(II) and remove it while MerPT actively transport Hg(II) into the cell, leading to a toxic bioaccumulation that inhibits bacterial growth.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1201
Illegal NgoMIV site found at 1249
Illegal NgoMIV site found at 1311
Illegal NgoMIV site found at 1522 - 1000COMPATIBLE WITH RFC[1000]