Difference between revisions of "Part:BBa K823034"
| (16 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K823034 short</partinfo> | <partinfo>BBa_K823034 short</partinfo> | ||
| − | 3x | + | 3x FLAG tag with RBS in [[Help:Assembly_standard_25|Freiburg standard]]. |
| + | |||
| + | Find out more about the design of our [https://static.igem.org/mediawiki/parts/b/b9/LMU-Munich_2012_Our_Freiburg_standard_FusionPrefix.pdf prefix with ribosome binding site]. | ||
prefix:GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC | prefix:GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC | ||
| Line 8: | Line 10: | ||
suffix:ACCGGTTAATACTAGTAGCGGCCGCTGCAGT | suffix:ACCGGTTAATACTAGTAGCGGCCGCTGCAGT | ||
| − | < | + | <p align="justify"> |
| + | The Flag-tag was the first epitope tag to be published ([http://www.nature.com/nbt/journal/v6/n10/full/nbt1088-1204.html T.P. Hopp, K.S. Prickett et al. (1988)]). It consists of eight hydrophobic aminoacids: DYKDDDDK and the 3x Flag tag is: <b>DYKDHDGDYKDHDIDYKDDDDK</b>. There are a variety of monoclonal antibodies against this tag, N-terminal as well as position insensitive.</p> | ||
| + | |||
| + | |||
| + | This is a part created by the LMU-Munich 2012 team. We added five tags to the registry, all in the Freiburg standard for N-and C-terminal fusions: | ||
| + | |||
| + | *[[Part:BBa_K823034|3x Flag - tag]] | ||
| + | |||
| + | *[[Part:BBa_K823035|HA - tag]] | ||
| + | |||
| + | *[[Part:BBa_K823036|cMyc - tag]] | ||
| + | |||
| + | *[[Part:BBa_K823037|His - tag]] | ||
| + | |||
| + | *[[Part:BBa_K823038| Strep - tag]] | ||
| + | |||
| + | |||
| + | |||
| + | Visit our project page for more usefull parts of our [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks '''''BacillusB'''''io'''B'''rick'''B'''ox]. This part was also evaluated in the publication [http://www.jbioleng.org/content/7/1/29 The ''Bacillus'' BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with ''Bacillus subtilis''] by Radeck ''et al.''. | ||
| + | |||
| + | ===Evaluation=== | ||
| + | <p align="justify"> | ||
| + | All 5 epitope tags were fused C- and N-terminally to GFP using the NgoMIV and AgeI restriction sites. These constructs were expressed in ''Bacillus subtils'' using [https://parts.igem.org/Part:BBa_K823026 pSB<sub>Bs</sub>0K-P<sub>spac</sub>]. This vector did not need to be induced by IPTG due to a premature stop codon in the lacI gene. | ||
| + | |||
| + | |||
| + | </p> | ||
| + | |||
| + | {| style="color:black;" cellpadding="3" width="70%" cellspacing="0" border="0" align="center" style="text-align:left;" | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| | ||
| + | |[[Image:LMU-Western_Blot_Tags.png|400px|center]] | ||
| + | |- | ||
| + | | style="width: 70%;background-color: #EBFCE4;" | | ||
| + | {| style="color:black;" cellpadding="0" width="100%" cellspacing="0" border="0" align="center" style="text-align:center;" | ||
| + | |style="width: 70%;background-color: #EBFCE4;" | | ||
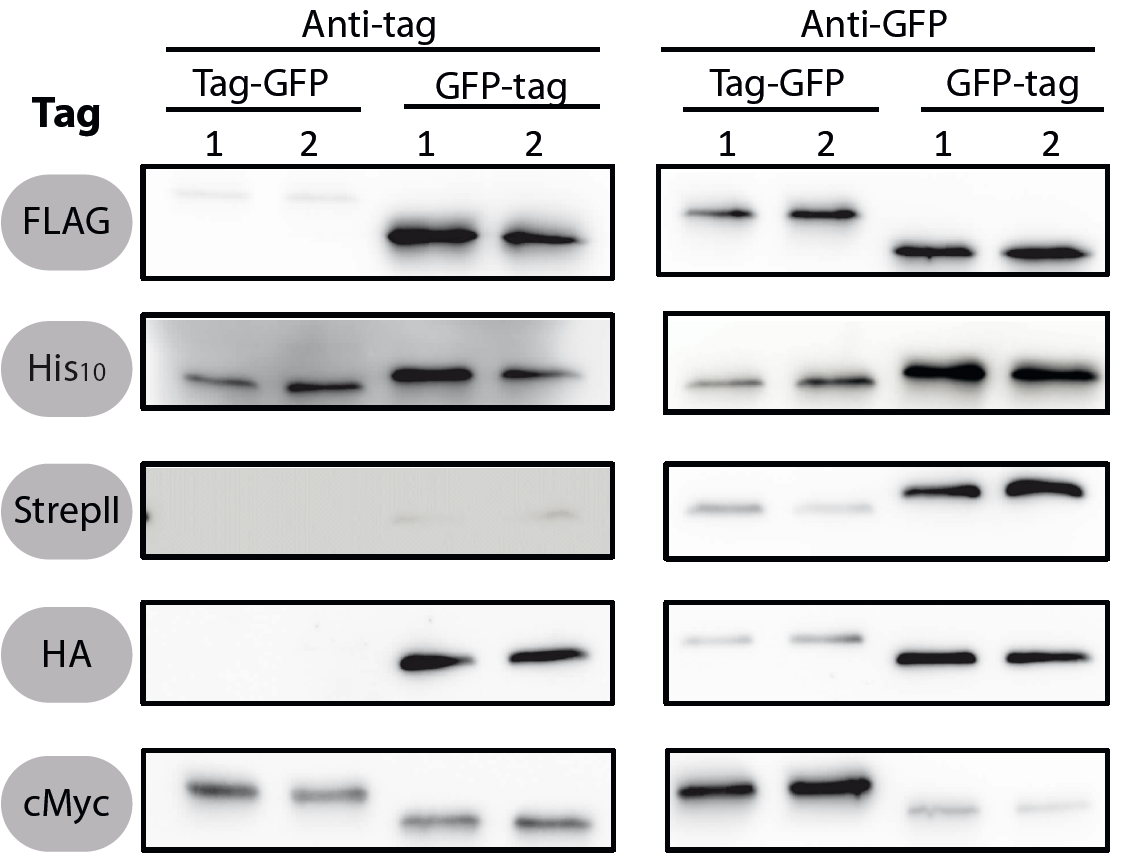
| + | <font color="#000000"; size="2"><p align="justify"> Fig. 1: Western blots of N- and C-terminal fusions of each tag to GFP, using the strains TMB1920 (Flag-gfp), TMB1921 (gfp-Flag), TMB1922 (HA-gfp), TMB1923 (gfp-HA), TMB1924 (cMyc-gfp), TMB1925 (gfp-cMyc), TMB1926 (His-gfp), TMB1927 (gfp-His), TMB1928 (StrepII-gfp) and TMB1929 (gfp-StrepII). For each construct, two independent clones were tested with epitope tag- and GFP-specific antibodies as a positive control. | ||
| + | |||
| + | |} | ||
| + | |} | ||
| + | |} | ||
| + | |||
| + | ===Methods=== | ||
| + | |||
| + | |||
| + | To verify the functionality of the epitope tags, Western blot analyses of the strains TMB1920-TMB1929 were performed. LB medium (15 ml) was inoculated 1:100 from overnight culture and grown at 37°C and 200 rpm to OD600 ~ 0.5. Of this, 10 ml were harvested by centrifugation (8000 × g, 5 min) and the pellets stored at -20°C. Pellets were resuspended in 1 ml disruption buffer (50 mM Tris–HCl pH 7.5, 100 mM NaCl) and lysed by sonication. Samples (12 μl of lysate) were loaded per lane on two 12.5% SDS-polyacrylamide gels and SDS-PAGE was performed according standard procedure [60]. One gel was stained with colloidal coomassie, the other one was used for protein transfer to a PVDF membrane (Merck Millipore, Billerica, MA, USA) by submerged blotting procedure (Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad, Hercules, CA, USA)). After protein transfer, the membranes were treated with the following antibodies and conditions. Detailed protocols can be found [http://www.jbioleng.org/content/7/1/29/suppl/S3 here]. | ||
| + | |||
| + | |||
| + | ''GFP'' | ||
| + | |||
| + | Probing with primary antibodies takes place with rabbit anti-GFP antibodies (1:3000, Epitomics, No. 1533). Horseradish-peroxidase (HRP)-conjugated anti-rabbit antibodies (1:2000, Promega, W401B) were used as secondary antibody. Hybridization of both antibodies was carried out in Blotto-buffer (2.5% (w/v) skim milk powder, 1 × TBS (50 mM Tris–HCl pH 7.6, 0.15 M NaCl)). | ||
| + | |||
| + | |||
| + | ''FLAG'' | ||
| + | |||
| + | Rabbit anti-FLAG (1:2000, Sigma, Anti-Flag polyclonal, F7425) and anti-rabbit-HRP (1:2000, Promega, W401B) were used in Blotto-buffer. | ||
| + | |||
| + | |||
| + | Chemiluminescence signals were detected after addition of the HRP-substrate Ace Glow (Peqlab, Erlangen, Germany) using a Fusion<sup>TM</sup> imaging system (Peqlab). | ||
| + | </p> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
Latest revision as of 17:05, 3 February 2014
3x FLAG tag (Freiburg standard+RBS)
3x FLAG tag with RBS in Freiburg standard.
Find out more about the design of our prefix with ribosome binding site.
prefix:GAATTCCGCGGCCGCTTCTAGATAAGGAGGAACTACTATGGCCGGC
suffix:ACCGGTTAATACTAGTAGCGGCCGCTGCAGT
The Flag-tag was the first epitope tag to be published ([http://www.nature.com/nbt/journal/v6/n10/full/nbt1088-1204.html T.P. Hopp, K.S. Prickett et al. (1988)]). It consists of eight hydrophobic aminoacids: DYKDDDDK and the 3x Flag tag is: DYKDHDGDYKDHDIDYKDDDDK. There are a variety of monoclonal antibodies against this tag, N-terminal as well as position insensitive.
This is a part created by the LMU-Munich 2012 team. We added five tags to the registry, all in the Freiburg standard for N-and C-terminal fusions:
- 3x Flag - tag
Visit our project page for more usefull parts of our [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks BacillusBioBrickBox]. This part was also evaluated in the publication [http://www.jbioleng.org/content/7/1/29 The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis] by Radeck et al..
Evaluation
All 5 epitope tags were fused C- and N-terminally to GFP using the NgoMIV and AgeI restriction sites. These constructs were expressed in Bacillus subtils using pSBBs0K-Pspac. This vector did not need to be induced by IPTG due to a premature stop codon in the lacI gene.
|
Methods
To verify the functionality of the epitope tags, Western blot analyses of the strains TMB1920-TMB1929 were performed. LB medium (15 ml) was inoculated 1:100 from overnight culture and grown at 37°C and 200 rpm to OD600 ~ 0.5. Of this, 10 ml were harvested by centrifugation (8000 × g, 5 min) and the pellets stored at -20°C. Pellets were resuspended in 1 ml disruption buffer (50 mM Tris–HCl pH 7.5, 100 mM NaCl) and lysed by sonication. Samples (12 μl of lysate) were loaded per lane on two 12.5% SDS-polyacrylamide gels and SDS-PAGE was performed according standard procedure [60]. One gel was stained with colloidal coomassie, the other one was used for protein transfer to a PVDF membrane (Merck Millipore, Billerica, MA, USA) by submerged blotting procedure (Mini Trans-Blot Electrophoretic Transfer Cell (Bio-Rad, Hercules, CA, USA)). After protein transfer, the membranes were treated with the following antibodies and conditions. Detailed protocols can be found [http://www.jbioleng.org/content/7/1/29/suppl/S3 here].
GFP
Probing with primary antibodies takes place with rabbit anti-GFP antibodies (1:3000, Epitomics, No. 1533). Horseradish-peroxidase (HRP)-conjugated anti-rabbit antibodies (1:2000, Promega, W401B) were used as secondary antibody. Hybridization of both antibodies was carried out in Blotto-buffer (2.5% (w/v) skim milk powder, 1 × TBS (50 mM Tris–HCl pH 7.6, 0.15 M NaCl)).
FLAG
Rabbit anti-FLAG (1:2000, Sigma, Anti-Flag polyclonal, F7425) and anti-rabbit-HRP (1:2000, Promega, W401B) were used in Blotto-buffer.
Chemiluminescence signals were detected after addition of the HRP-substrate Ace Glow (Peqlab, Erlangen, Germany) using a FusionTM imaging system (Peqlab).
Usage and Biology
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]