Difference between revisions of "Part:BBa K1189029"
| (5 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
| − | |||
<html> | <html> | ||
| − | E/K coils are synthetic coiled-coil domains designed specifically to bind to each other with high affinity and specificity (Litowski and Hodges, 2002) (Figure 1). They are composed of a heptad repeat that forms a coil structures that are able to interact with each other. These coils are able to interact with each other in an anti-parallel fashion that makes them useful for applications such as peptide capture, protein purification and in biosensors. For our project we decided to make use of the IAAL E3/K3 coils (BBa_K118901, BBa_K1189011) due to the balance they offer between affinity and specificity (Table 1). | + | <h1>TALE A fused to C-terminus K coil with his tag</h1> |
| + | |||
| + | <p>For information on TALEA refer to <a href="https://parts.igem.org/Part:BBa_K1189022"><b>BBa_K1189022</b></a>.</p> | ||
| + | |||
| + | <p> | ||
| + | E/K coils are synthetic coiled-coil domains designed specifically to bind to each other with high affinity and specificity (Litowski and Hodges, 2002) (Figure 1). They are composed of a heptad repeat that forms a coil structures that are able to interact with each other. These coils are able to interact with each other in an anti-parallel fashion that makes them useful for applications such as peptide capture, protein purification and in biosensors. For our project we decided to make use of the IAAL E3/K3 coils (BBa_K118901, BBa_K1189011) due to the balance they offer between affinity and specificity (Table 1). </p> | ||
<figure> | <figure> | ||
<img src=" https://static.igem.org/mediawiki/parts/8/87/K_coilHis-TALE_APlacI.png" width="170" height="80"> | <img src=" https://static.igem.org/mediawiki/parts/8/87/K_coilHis-TALE_APlacI.png" width="170" height="80"> | ||
| − | + | <figcaption> | |
| + | <p><b>Figure 1.</b> Optimized TALE A with his tag and a C-terminus K coil.</p> | ||
| + | </figcaption> | ||
</figure> | </figure> | ||
| + | |||
| + | <br></br> | ||
| + | |||
<figure> | <figure> | ||
<img src="https://static.igem.org/mediawiki/2013/a/a9/UCalgary2013TRCoilrenderpng.png" alt="Coiled-coils" width="193" height="300"> | <img src="https://static.igem.org/mediawiki/2013/a/a9/UCalgary2013TRCoilrenderpng.png" alt="Coiled-coils" width="193" height="300"> | ||
<figcaption> | <figcaption> | ||
| − | <p><b>Figure | + | <p><b>Figure 2.</b> Ribbon visualization of the E3/K3 IAAL coiled-coils.</p> |
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| + | |||
<center><b>Table 1. Coil Peptide Sequences</b></center> | <center><b>Table 1. Coil Peptide Sequences</b></center> | ||
<center><table width="400" border="1"> | <center><table width="400" border="1"> | ||
| Line 55: | Line 64: | ||
</p> | </p> | ||
</figcaption> | </figcaption> | ||
| − | </figure> </html> | + | </figure> |
| + | |||
| + | <p> | ||
| + | We ordered 60mer FAM-labeled [A] (TALE A target sequence) and hybridized them with their reverse complement oligo to make double stranded pieces of DNA containing the target sequence of our TALEs. Using these target sequences and following the <a href="http://2013.igem.org/Team:Calgary/Notebook/Protocols/FunctionalityAssayOnNitrocellulose" >TALE Nitrocellulose Functionality Assay</a>, we showed that TALEA binds to its target sequence. We incubated Ferritin fused to an Ecoil (<a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189018" >BBa_K1189018</a>) to TALE fused to a Kcoil (<a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189029" >BBa_K1189029</a>) to make the FerriTALE complex. The complex was then blotted on strips of nitrocellulose paper. The strips were then blocked with milk and soaked in the appropriate DNA solution. Finally, the strips were washed and imaged (figure 14 and 15). We performed a densitometery test on these results and were able to calculate the dissociation constant of the TALEs. | ||
| + | </p> | ||
| + | |||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/9/95/Calary2013TALEABlotWithKinetics.png" width="800" height="346"> | ||
| + | <figcaption> | ||
| + | <p><b>Figure 5.</b> (A) Dot blot of FerriTALE A exposed to FAM labeled DNA containing the [A] TALE A target sequence (<a href="http://2013.igem.org/Team:Calgary/Notebook/Protocols/FunctionalityAssayOnNitrocellulose" >protocol</a>). 1.5µg of TALEA+Kcoil and 1µg of ferritin with E coil were incubated for 1 hour to make the FerriTALE complex and the complex was blotted on a strip. The blots were then exposed to 1.66 mM FAM labeled [A] TALE A target site from 1 to 90 minutes as indicated on the strips. "x" is a FerriTALE that was exposed to FAM labeled DNA prior to being blotted onto the nitrocellulose. The kinetics from the densitometry is shown in section B of the figure. The Kd from this plot was determined to be 293nM.</p> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | |||
| + | |||
| + | <p>We also wanted to show that our TALEs are specific to their target sequence. So we did another experiment to test whether TALEA fused to a Kcoil (<a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189029" >BBa_K1189029</a>) can bind to the TALE B target site ([B]). This experiment showed that TALEA only binds to [A] and not [B]. We showed not only that <span class="Yellow"><b>TALEs bind DNA</b></span> , they are also <span class="Yellow"><b>specific</b></span> to their own target site (Figure 6). | ||
| + | </p> | ||
| + | |||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/5/5c/Ucalgary_2013_ocotber._TALE_specificity.png" width="363" height="334"> | ||
| + | <figcaption> | ||
| + | <p><b>Figure 6.</b> (A) A Dot blot of TALE A on nitrocellulose paper (<a href="http://2013.igem.org/Team:Calgary/Notebook/Protocols/FunctionalityAssayOnNitrocellulose" >protocol</a>). A6 is TALE A soaked in 1.66mM FAM labeled [B] TALE B target sequence. A7 is TALE A soaked in 1.66mM FAM labeled [A] TALE A target sequence. A2 is TALE A soaked in 1mM FAM labeled [B] TALE B target sequence. A3 is TALE A soaked in 1mM FAM labeled [A] TALE A target sequence. On A- strip no protein was blotted and it was soaked in 1.66mM [A]. All strips were soaked in DNA solution for 90 minutes. (B) 1µL of the DNA solutions used for soaking were blotted on nitrocellulose and a picture was taken instantly, to indicate that both [A] and [B] fluoresce to the same extent. All the DNA solutions contained 1900ng/uL salmon sperm DNA as a competitor for the TALE target site. | ||
| + | |||
| + | </figcaption> | ||
| + | </figure> | ||
| + | |||
| + | |||
| + | </html> | ||
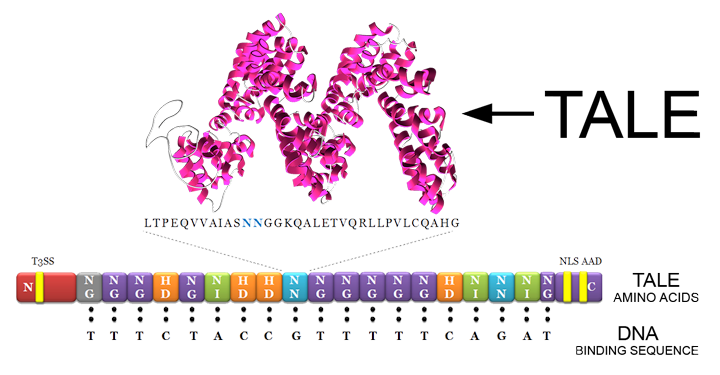
[[File:Calgary2013-TALE-RVD.png]] | [[File:Calgary2013-TALE-RVD.png]] | ||
Latest revision as of 07:49, 2 November 2013
TALE A fused to C-terminus K coil with his tag
For information on TALEA refer to BBa_K1189022.
E/K coils are synthetic coiled-coil domains designed specifically to bind to each other with high affinity and specificity (Litowski and Hodges, 2002) (Figure 1). They are composed of a heptad repeat that forms a coil structures that are able to interact with each other. These coils are able to interact with each other in an anti-parallel fashion that makes them useful for applications such as peptide capture, protein purification and in biosensors. For our project we decided to make use of the IAAL E3/K3 coils (BBa_K118901, BBa_K1189011) due to the balance they offer between affinity and specificity (Table 1).

Figure 1. Optimized TALE A with his tag and a C-terminus K coil.

Figure 2. Ribbon visualization of the E3/K3 IAAL coiled-coils.
| Coil Name | Peptide Sequence |
| IAAL E3 | NH2-EIAALEKEIAALEKEIAALEK-COOH |
| IAAL K3 | NH2-KIAALKEKIAALKEKIAALKE-COOH |
These E3/K3 coils are able to form heterodimers due to the hydrophobic residues contained within the heptad repeat. In our case these are isoleucine and leucine residues. Designated by empty arrows in the helical wheel diagram below (Figure 2) these residues form the core of the binding domain of the coils. In order to prevent the homodimerization of these coils charged residues are included in the design. The electrostatic interactions between glutamic acid and lysine residues prevent an E-coil from binding with an E-coil for example. These parts were already in the registry, however the DNA was never received, so we built, sequenced and re-submitted them.

Figure 2. A helical wheel representation of the IAAL E3/K3 coiled-coil heterodimer viewed as a cross-section based off of a similar figure created by Litowski and Hodges (2002). The peptide chain propagates into the page from the N- to the C- terminus. Hydrophobic interactions between the coils are indicated by the clear wide arrows. The intermolecular electrostatic interactions between the coils are displayed by the thin curved arrow (eg. Between Glu15 and Lys20) Letters a, b, c,and d designate the positions of IAAL repeat in the heptapeptide. The e and g positions are occupied by the charged residues.
We evaluated the binding of our coils using other constructs that make use of the E and K coil parts submitted. In the case of the coils we were interested to see if the K-coil fused to TALE proteins (BBa_K1189029, BBa_K1189030) could bind to the E-coil found on one of our Prussian blue ferritin constructs (BBa_K1189018). To complete this task we placed the TALE on the membrane, washed and blocked the membrane. The ferritin protein with the complimentary coil was then added to the membrane. If this coil successfully binds to the other coil then the ferritin will not be washed off during the next wash step. We can then see if Prussian blue ferritin is bound by adding a TMB substrate solution that will cause a colour change. To this extent we saw a blue ring in this trial indicating a positive result. This suggests that our coils are actually binding in an in vitro system.
Another interesting element of this assay is why we used two variants of the TALE K-coil negative control. A blue ring on our TALE negative control confirmed our fear that during the second protein application and wash step that some of the ferritin with coil proteins would drift over and bind to the TALE K-coils on the nitrocellulose. This did not occur for our separate negative control (Figure 3).

Figure 3. This basic qualitative assay was used to inform us whether certain elements of our system are able to bind to each other. Our TALE proteins were mounted to the membrane along with positive controls of three Prussian blue variants; two recombinant ferritins and one commercial protein. The membranes were then washed and blocked. Prussian blue ferritin with a coil was added to our TALE protein containing a coil. Prussian blue ferritin with a TALE that could bind to the DNA held by another TALE on the membrane was also added. A TMB substrate solution was added to cause a colourimetric change over 5 minutes. Positive results are indicated by dark rings of colour. Negative controls include a TALE with a coil on the same membrane and the same TALE and bovine serum albumin on separate membranes that were treated separately. Image contrast was altered to make the results more clear on a digital monitor; the same changes were applied to each element of the figure.
We also performed an immunoprecipitation assay to demonstrate the binding of the E/K coils (Figure 4).

Figure 4. Assay showing coiled-coil interaction in vitro. Crude lysates from a negative control (RFP), GFP-Ecoil and His-Kcoil were combined together to investigate interaction and immunoprecipitated with GFP or an isotype control and then further probed with α-His antibody. Only in the presence of both GFP and a His tag we see a band indicating interaction.
We ordered 60mer FAM-labeled [A] (TALE A target sequence) and hybridized them with their reverse complement oligo to make double stranded pieces of DNA containing the target sequence of our TALEs. Using these target sequences and following the TALE Nitrocellulose Functionality Assay, we showed that TALEA binds to its target sequence. We incubated Ferritin fused to an Ecoil (BBa_K1189018) to TALE fused to a Kcoil (BBa_K1189029) to make the FerriTALE complex. The complex was then blotted on strips of nitrocellulose paper. The strips were then blocked with milk and soaked in the appropriate DNA solution. Finally, the strips were washed and imaged (figure 14 and 15). We performed a densitometery test on these results and were able to calculate the dissociation constant of the TALEs.

Figure 5. (A) Dot blot of FerriTALE A exposed to FAM labeled DNA containing the [A] TALE A target sequence (protocol). 1.5µg of TALEA+Kcoil and 1µg of ferritin with E coil were incubated for 1 hour to make the FerriTALE complex and the complex was blotted on a strip. The blots were then exposed to 1.66 mM FAM labeled [A] TALE A target site from 1 to 90 minutes as indicated on the strips. "x" is a FerriTALE that was exposed to FAM labeled DNA prior to being blotted onto the nitrocellulose. The kinetics from the densitometry is shown in section B of the figure. The Kd from this plot was determined to be 293nM.
We also wanted to show that our TALEs are specific to their target sequence. So we did another experiment to test whether TALEA fused to a Kcoil (BBa_K1189029) can bind to the TALE B target site ([B]). This experiment showed that TALEA only binds to [A] and not [B]. We showed not only that TALEs bind DNA , they are also specific to their own target site (Figure 6).

Figure 6. (A) A Dot blot of TALE A on nitrocellulose paper (protocol). A6 is TALE A soaked in 1.66mM FAM labeled [B] TALE B target sequence. A7 is TALE A soaked in 1.66mM FAM labeled [A] TALE A target sequence. A2 is TALE A soaked in 1mM FAM labeled [B] TALE B target sequence. A3 is TALE A soaked in 1mM FAM labeled [A] TALE A target sequence. On A- strip no protein was blotted and it was soaked in 1.66mM [A]. All strips were soaked in DNA solution for 90 minutes. (B) 1µL of the DNA solutions used for soaking were blotted on nitrocellulose and a picture was taken instantly, to indicate that both [A] and [B] fluoresce to the same extent. All the DNA solutions contained 1900ng/uL salmon sperm DNA as a competitor for the TALE target site.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1922
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 2651
Illegal AgeI site found at 677
Illegal AgeI site found at 1258 - 1000COMPATIBLE WITH RFC[1000]