Difference between revisions of "Part:BBa K1172905"
| Line 15: | Line 15: | ||
For more details about the function of the Biosafety-System in general, click [http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System here]</p><br> | For more details about the function of the Biosafety-System in general, click [http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System here]</p><br> | ||
| − | Moreover the BioBrick is not limited to this Biosafety-System, as it can be used for the regulated trancription of any coding sequence behind the P<sub>''BAD''</sub> promoter, opening the possibility of an antibiotic-free selection in the [http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_Strain Biosafety-Strain]. | + | Moreover the BioBrick is not limited to this Biosafety-System, as it can be used for the regulated trancription of any coding sequence behind the P<sub>''BAD''</sub> promoter, opening the possibility of an antibiotic-free selection in the [http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_Strain Biosafety-Strain].</p><br> |
| − | <span class='h3bb'>Sequence and Features</span> | + | <span class='h3bb'>'''Sequence and Features'''</span> |
<partinfo>BBa_K1172905 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1172905 SequenceAndFeatures</partinfo> | ||
| Line 26: | Line 26: | ||
<!-- --> | <!-- --> | ||
| − | ===''' | + | ==='''Biosafety-System araCtive'''=== |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
<br> | <br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<p align="justify"> | <p align="justify"> | ||
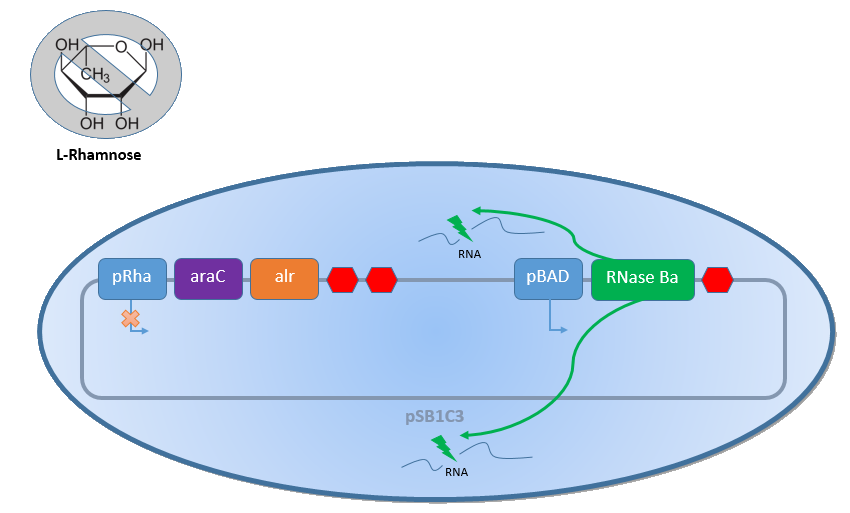
| − | + | Combining the [http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_S genes] described above with the Biosafety-Strain K-12 ∆''alr'' ∆''dadX'' results in a powerful device, allowing us to control the bacterial cell division. The control of the bacterial growth is possible either active or passive. Active by inducing the P<sub>''BAD''</sub> promoter with L-arabinose and passive by the induction of L-rhamnose. The passive control makes it possible to control the bacterial cell division in a defined closed environment, like the MFC, by continuously adding L-rhamnose to the medium. As shown in the Figure 1 below, this leads to an expression of the essential alanine racemase (''alr'') and the AraC regulator, so that the expression of the RNase Ba is repressed. </p><br> | |
| − | |||
| − | + | [[File:IGEM Bielefeld 2013 Biosafety System S+ 2.png|600px|thumb|center|'''Figure 1:''' Biosafety-System araCtive in the presence of L-rhamnose. The essential alanine racemase (Alr) and the repressor AraC are expressed, resulting in a repression of the expression of the RNAse Ba. Consequently the bacteria show normal growth behaviour.]] | |
| − | [[File: | + | |
| − | + | ||
| − | |||
<br> | <br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<p align="justify"> | <p align="justify"> | ||
| − | + | In the event that bacteria exit the defined environment of the MFC or L-rhamnose is not added to the medium any more, both the expression of the alanine racemase (Alr) and the AraC regulatordecrease, so that the expression of the toxic RNase Ba (Barnase) begins. The cleavage of the intracellular RNA by the Barnase and the lack of synthesized D-alanine, caused by the repressed alanine racemase inhibit the cell division and makes sure that the bacteria can only grow in the defined environment or the device of choice respectively. </p><br> | |
| − | + | [[File:IGEM Bielefeld 2013 Biosafety System S ohne Rhamnose 2.png|600px|thumb|center|'''Figure 2:''' Active Biosafety-System araCtive outside of a defined environment lacking L-rhamnose. Both the expression of the alanine racemase (Alr) and AraC repressor are reduced and ideally completely shutdown. In contrast, the expression of the RNase ba (Barnase) is turned on, leading to cell death by RNA cleavage.]] | |
| − | |||
| − | |||
| − | |||
<br> | <br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 15:23, 30 October 2013
Part 1 of the Biosafety-System araCtive
Usage and Biology
These biobrick is the first part of the Biosafety-System AraCtive BBa_K1172909, which is an improvement of the Biobrick BBa_K914014 by replacing:
- the first promoter arabinose PBAD by the rhamnose promoter PRha to obtain a lower basal transcription.
- intgration of the alanine racemase BBa_K1172901(alr) for gaining higher plasmid stability and for taking advantage of the double-kill switch mechanism.
- the repressor LacI BBa_C0012 to AraC and the lac promoter to the arabinose promoter PBAD for a higher repression and lower basal transcription of the second part.
For more details about the seperate genes in this Biosafety-System and their function, click [http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_S here]
For more details about the function of the Biosafety-System in general, click [http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System here]</p>
Moreover the BioBrick is not limited to this Biosafety-System, as it can be used for the regulated trancription of any coding sequence behind the PBAD promoter, opening the possibility of an antibiotic-free selection in the [http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_Strain Biosafety-Strain].</p>
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1374
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1298
Illegal BamHI site found at 2000 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 1416
Illegal AgeI site found at 1716 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1173
Biosafety-System araCtive
Combining the [http://2013.igem.org/Team:Bielefeld-Germany/Biosafety/Biosafety_System_S genes] described above with the Biosafety-Strain K-12 ∆alr ∆dadX results in a powerful device, allowing us to control the bacterial cell division. The control of the bacterial growth is possible either active or passive. Active by inducing the PBAD promoter with L-arabinose and passive by the induction of L-rhamnose. The passive control makes it possible to control the bacterial cell division in a defined closed environment, like the MFC, by continuously adding L-rhamnose to the medium. As shown in the Figure 1 below, this leads to an expression of the essential alanine racemase (alr) and the AraC regulator, so that the expression of the RNase Ba is repressed.
In the event that bacteria exit the defined environment of the MFC or L-rhamnose is not added to the medium any more, both the expression of the alanine racemase (Alr) and the AraC regulatordecrease, so that the expression of the toxic RNase Ba (Barnase) begins. The cleavage of the intracellular RNA by the Barnase and the lack of synthesized D-alanine, caused by the repressed alanine racemase inhibit the cell division and makes sure that the bacteria can only grow in the defined environment or the device of choice respectively.
References
- Baldoma L and Aguilar J (1988) Metabolism of L-Fucose and L-Rhamnose in Escherichia coli: Aerobic-Anaerobic Regulation of L-Lactaldehyde Dissimilation [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC210658/pdf/jbacter00179-0434.pdf|Journal of Bacteriology 170: 416 - 421.].
- Carafa, Yves d'Aubenton Brody, Edward and Claude (1990) Thermest Prediction of Rho-independent Escherichia coli Transcription Terminators - A Statistical Analysis of their RNA Stem-Loop Structures [http://ac.els-cdn.com/S0022283699800059/1-s2.0-S0022283699800059-main.pdf?_tid=ede07e2a-2a92-11e3-b889-00000aab0f6c&acdnat=1380629809_2d1a59e395fc69c8608ab8b5aea842f7|Journal of molecular biology 216: 835 - 858].
- Cass, Laura G. and Wilcoy, Gary (1988) Novel Activation of araC Expression and a DNA Site Required for araC Autoregulation in Escherichia coli B/r [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC211425/pdf/jbacter00187-0394.pdf|Journal of Bacteriology 170: 4174 - 4180].
- Hamilton, Eileen P. and Lee, Nancy (1988) Three binding sites for AraC protein are required for autoregulation of araC in Escherichia coli [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC279856/pdf/pnas00258-0029.pdf|Proc Natl Acad Sci U S A 85: 1749 - 53].
- Mossakowska, Danuta E. Nyberg, Kerstin and Fersht, Alan R. (1989) Kinetic Characterization of the Recombinant Ribonuclease from Bacillus amyloliquefaciens (Barnase) and Investigation of Key Residues in Catalysis by Site-Directed Mutagenesis [http://pubs.acs.org/doi/pdf/10.1021/bi00435a033|Biochemistry 28: 3843 - 3850.].
- Paddon, C. J. Vasantha, N. and Hartley, R. W. (1989) Translation and Processing of Bacillus amyloliquefaciens Extracellular Rnase [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC209718/pdf/jbacter00168-0575.pdf|Journal of Bacteriology 171: 1185 - 1187.].
- Schleif, Robert (2010) AraC protein, regulation of the L-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action [http://gene.bio.jhu.edu/Ourspdf/127.pdf|FEMS microbial reviews 34: 779 - 796.].
- Voss, Carsten Lindau, Dennis and Flaschel, Erwin (2006) Production of Recombinant RNase Ba and Its Application in Downstream Processing of Plasmid DNA for Pharmaceutical Use [http://onlinelibrary.wiley.com/doi/10.1021/bp050417e/pdf|Biotechnology Progress 22: 737 - 744.].
- Walsh, Christopher (1989) Enzymes in the D-alanine branch of bacterial cell wall peptidoglycan assembly. [http://www.jbc.org/content/264/5/2393.long|Journal of biological chemistry 264: 2393 - 2396.]
- Wickstrum, J.R., Santangelo, T.J., and Egan, S.M. (2005) Cyclic AMP receptor protein and RhaR synergistically activate transcription from the L-rhamnose-responsive rhaSR promoter in Escherichia coli. [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1251584/?report=reader|Journal of Bacteriology 187: 6708 – 6719.].

