Difference between revisions of "Part:BBa K1189031:Experience"
| Line 5: | Line 5: | ||
===Applications of BBa_K1189031=== | ===Applications of BBa_K1189031=== | ||
<html> | <html> | ||
| + | <p>For information on TALEA refer to <a href="https://parts.igem.org/Part:BBa_K1189022"><b>BBa_K1189022</b></a>.</p> | ||
| + | |||
| + | <p>This part was built to function as both our detector (TALE A) and our reporter (beta-lactamase). The part was built with the lacI IPTG inducible promoter J04500, with RBS, and it has a His-tag for protein purification.</p> | ||
| + | |||
| + | <p>TALE A was inspired by the award winning <a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K782004">TALE A</a> from the award winning <a href="http://2012.igem.org/Team:Slovenia">2012 Slovenian iGEM project</a>. The <a href="http://2013.igem.org/Team:Calgary">iGEM Calgary 2013</a> used this TALE and its associated DNA binding sequence to build a proof of concept TALE based DNA detector. In the case of <a href="https://parts.igem.org/Part:BBa_K1189031">BBa_K1189031</a>, the Calgary team used beta-lactamase as a reporter enzyme to indicate when the TALE is bound to DNA. </p> | ||
| + | |||
<figure> | <figure> | ||
| − | <img src="https://static.igem.org/mediawiki/ | + | <img src="https://static.igem.org/mediawiki/parts/5/51/YYC_2013_PlacI_%2B_His_%2B_TALE_%2B_Blac.jpg"> |
| + | </figure> | ||
| + | </html> | ||
| + | ===Applications of BBa_K1189031=== | ||
| + | <html> | ||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/0/03/YYC2013_Blac_Amp_Survival_Assay_with_colonies.jpg"></img> | ||
<figcaption> | <figcaption> | ||
<p><b>Figure 1. </b>Absorbance values at 600nm for each tube at four different time points: 0, 30, 60 and 120min. The cultures that expressed beta-lactamase (<a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189007"> | <p><b>Figure 1. </b>Absorbance values at 600nm for each tube at four different time points: 0, 30, 60 and 120min. The cultures that expressed beta-lactamase (<a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189007"> | ||
| Line 60: | Line 72: | ||
</figure> | </figure> | ||
<figure> | <figure> | ||
| − | <img src="https://static.igem.org/mediawiki/2013 | + | <img src="https://static.igem.org/mediawiki/2013/3/38/YYC2013_Blac_Amp_Survival_Assay_with_protein_24h.jpg"> |
<figcaption> | <figcaption> | ||
<p><b>Figure 3. </b>Absorbance values at 600nm after 24h. Amounts from 0.1µg to 20µg of TALE A-link-Beta-lactamase (<a href=" https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031"> | <p><b>Figure 3. </b>Absorbance values at 600nm after 24h. Amounts from 0.1µg to 20µg of TALE A-link-Beta-lactamase (<a href=" https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031"> | ||
| Line 70: | Line 82: | ||
</figure> | </figure> | ||
<figure> | <figure> | ||
| − | <img src="https://static.igem.org/mediawiki/2013 | + | <img src="https://static.igem.org/mediawiki/2013/d/de/YYC2013_Blac_Amp_Survival_Assay_with_protein_3_time_points.jpg"> |
<figcaption> | <figcaption> | ||
<p><b>Figure 4. </b>Absorbance values at 600nm in different time points. Amounts from 1.0µg to 10µg of TALE A-link-Beta-lactamase (<a href=" https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031"> | <p><b>Figure 4. </b>Absorbance values at 600nm in different time points. Amounts from 1.0µg to 10µg of TALE A-link-Beta-lactamase (<a href=" https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031"> | ||
| Line 106: | Line 118: | ||
</figcaption> | </figcaption> | ||
</figure> | </figure> | ||
| + | </html> | ||
| + | |||
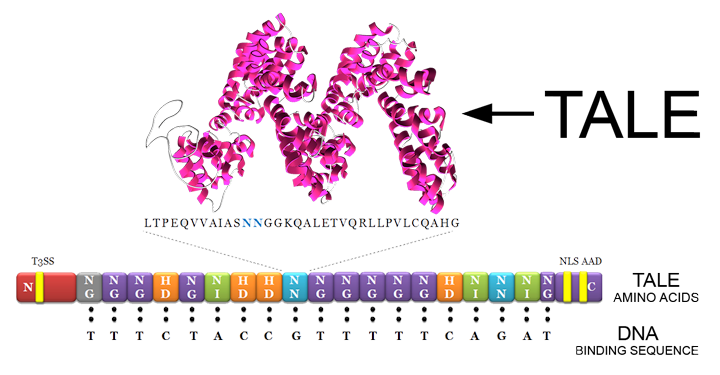
| + | [[File:Calgary2013-TALE-RVD.png]] | ||
| + | |||
| + | <html> | ||
| + | |||
| + | <p>Additionally, we have demonstrated the same pH change of benzylpenicillin to penicillinoic acid by the TALE A β-lactamase fusion (<a href=" https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031">BBa_K1189031</a>) with bromothymol blue. The pH change causes the bromothymol blue to change in colour from blue to yellow as it gets more acidic (Figure 11 & 12). In the presence of TALE A β-lactamase fusion we see the colour change to yellow whereas the negatives not containing TALE A β-lactamase fusion remains blue (Figure 11). We did a kinetic analysis at 616 nm every 30 seconds and as the blue colour disappears, the absorbance at 616 nm decreases (Figure 12). Therefore, the lower the decrease the better the TALE A β-lactamase fusion reporter activity. Our kinetic activity shows that the biggest decrease is in our positive recombinant β-lactamase followed by the TALE A β-lactamase fusion at 10 micrograms. This decrease is lessened as we decrease the amount of TALE A β-lactamase fusion. We can also show that in our negatives with no TALE A β-lactamase fusion we do not have a decrease in absorbance. We have demonstrated the reporter activity both qualitatively (Figure 10 & 11) and quantitatively (Figure 12). </p> | ||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/c/c7/UCalgary2013TRBetalactamasecolourpsd.png" alt="Beta-lactamase Visual Assay" width="432" height="599"> | ||
| + | <figcaption> | ||
| + | <p><b>Figure 11.</b>Change in pH catalyzed by TALE A linked to β-lactamase (<a href=" https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031">BBa_K1189031</a>) using benzylpenicillin. Bromothymol blue was used to keep track of this colour change. Absorbance readings were taken at 616 nm every 30 seconds. Different amounts of TALE A linked to β-lactamase (<a href=" https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031">BBa_K1189031</a>) were tested. Commercial β-lactamase was used as a positive control. Negative controls included were bovine serum albumin, β-lactamase without the substrate and the substrate by itself.</p> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | <p> In order to demonstrate that we can successfully capture target DNA with two TALEs we did a capture TALE assay. TALE B was incubated with DNA containing target sites for TALE A and TALE B and blotted on nitrocellulose. After blocking and washing, TALE A β-lactamase fusion (<a href=" https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031">BBa_K1189031</a>) was added to the nitrocellulose strips. The strips were transferred into a 96 well plate to which a benzylpenicillin substrate solution with phenol red was added. If the TALE A β-lactamase fusion bound to the target site for TALE A then the solution will change colour from pink to clear. If TALE A β-lactamase fusion was not present, the solution will remain pink. We can show that the first four samples which have TALE B with DNA for TALE A and TALE B show a colour change indicating that we are successfully capturing the target DNA and reporting it. Furthermore we can also show that when we add non-specific DNA we do not see a colour change demonstrating that we can successfully capture only specific DNA and report its presence with an easy visual colourimetric output. </p> | ||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/6/68/UCalgary2013TRBetalactamaseassay.png" alt="Beta-lactamase Catalytic Assay" width="800" height="442"> | ||
| + | <figcaption> | ||
| + | <p><b>Figure 12.</b>Change in pH catalyzed by TALE A linked to β-lactamase (<a href=" https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031">BBa_K1189031</a>) using benzylpenicillin. Bromothymol blue was used to keep track of this colour change. Absorbance readings were taken at 616 nm every 30 seconds. Different amounts of TALE A linked to β-lactamase (<a href=" https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031">BBa_K1189031</a>) were tested. Commercial β-lactamase was used as a positive control. Negative controls included were bovine serum albumin, β-lactamase without the substrate and the substrate by itself.</p> | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/5/54/TALE_B_and_TALE_A_B-lac_DNA_capture_assay.png" alt="TALE DNA Capture Assay" width="800" height="600"> | ||
| + | <figcaption> | ||
| + | |||
| + | |||
| + | |||
| + | <p><b>Figure 6: </b> TALE capture assay was done with TALE B (<a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189001"> | ||
| + | <span class="Green"><b> | ||
| + | BBa_K1189001 | ||
| + | </b></span> | ||
| + | </a >)and TALE A B-lac fusion (<a href=" https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031"> | ||
| + | <span class="Green"><b> | ||
| + | BBa_K1189031 | ||
| + | </b></span> | ||
| + | </a >) and DNA (<a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189006"> | ||
| + | <span class="Green"><b> | ||
| + | BBa_K1189006 | ||
| + | </b></span> </a >) | ||
| + | with both target sequences. If capture is successful, the B-lac is present in the well giving a colour change from pink to yellow when subjected to benzylpenicillin substrate solution within 20 minutes. The only wells that change colour are the first four wells which contain TALE B, specific DNA, and TALE A B-lac fusion and our positive control wells which are the controls for our fusion TALE A B-lac protein and our positive recombinant b-lac. All our other controls including our test using a non-specific sequence of DNA remained pink . This preliminary characterization data demonstrates that the TALEs are able to bind to DNA with specificity. Additionally it also shows that our system of capturing DNA with two detector TALEs and then subsequent reporting of the DNA’s presence works. | ||
| + | </figcaption> | ||
| + | </figure> | ||
| + | |||
| + | <p>This assay shows that we can <b> capture our target DNA </b> with two detector TALEs with <b>specificity </b>. Additionally, <b> we can report whether that DNA has been captured</b> and is present in the sample, which is a very important concept for our sensor system. </p> | ||
| + | <p>To conclude, we have demonstrated that we can build, express, and purify this part. This <b> biobrick has also been characterized to show both its ability to be a good reporter and its ability to be able to bind to DNA with specificity. </b> </p> | ||
| + | |||
| + | |||
| + | <p> | ||
| + | To further see if the TALE capture assay works (figure 17). In order to do this, we blotted TALEB fused to a K coil (<a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189030" >BBa_K1189030</a>) on a nitrocellulose strip. Subsequently the strips were blocked with 5% skimmed milk in TBST buffer. The strips were then washed and soaked in a solution containing [A] and [B] on the same plasmid(<a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189006" >BBa_K1189006</a>). The strips were then washed again with TALE binding buffer and soaked in a solution containing TALEA+β-lac (<a href=https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031 >BBa_K1189031</a>). Then a dot blot was done on the strips with β-lactamase primary antibody and an hrp conjugated secondary antibody. This assay demonstrated that the <span class="Yellow"><b>TALEs can capture DNA</b></span>. | ||
| + | </p> | ||
| + | <figure> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/4/4f/Calgary2013_Robert_Dual_TALEs_Densitometry_Assay.png" width="800" height="306"> | ||
| + | <figcaption> | ||
| + | <p><b>Figure 7.</b> Figure on the left: For the dual TALE lanes TALEB fused to a K coil (<a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189030" >BBa_K1189030</a>) was blotted on a nitrocellulose strip. Subsequently the strips were blocked with 5% skimmed milk in TBST buffer. The strips were then washed and soaked in a solution containing [A] and [B] on the same plasmid(<a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189006" >BBa_K1189006</a>). The strips were then washed again with TALE binding buffer and soaked in a solution containing TALEA+β-lac (<a href=https://parts.igem.org/wiki/index.php?title=Part:BBa_K1189031 >BBa_K1189031</a>). Then a blot dot was done on the strips with β-lactamase primary antibody and an hrp conjugated secondary antibody. In negative 1 the strip was not soaked in the DNA solution. In Negative 2, the strip was not soaked in TALEA+β-lac solution. For positive controls, TALEA+βlac was blotted on the strip at the beginning instead of TALEB+K coil. The figure on the right shows the result of the densitometery we performed on the dot blot assay. | ||
| + | |||
| + | </figcaption> | ||
| + | |||
</html> | </html> | ||
===User Reviews=== | ===User Reviews=== | ||
Revision as of 04:45, 29 October 2013
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K1189031
For information on TALEA refer to BBa_K1189022.
This part was built to function as both our detector (TALE A) and our reporter (beta-lactamase). The part was built with the lacI IPTG inducible promoter J04500, with RBS, and it has a His-tag for protein purification.
TALE A was inspired by the award winning TALE A from the award winning 2012 Slovenian iGEM project. The iGEM Calgary 2013 used this TALE and its associated DNA binding sequence to build a proof of concept TALE based DNA detector. In the case of BBa_K1189031, the Calgary team used beta-lactamase as a reporter enzyme to indicate when the TALE is bound to DNA.

Applications of BBa_K1189031
Figure 1. Absorbance values at 600nm for each tube at four different time points: 0, 30, 60 and 120min. The cultures that expressed beta-lactamase (
BBa_K1189007
) showed higher absorbance levels, showing that the cells were able to grow in the presence of ampicillin.
In addition to that, we have purified our beta-lactamase (
BBa_K1189007
) and our mobile TALE A linked to beta-lactamase construct (
BBa_K1189031
)
(Figure 2) and we have demonstrated that beta-lactamase retained its enzymatic activity for both proteins. We repeated a variation of ampicillin survival assay where we pretreated LB containing ampicillin and chloramphenicol with our purified TALE A linked to beta-lactamase (
BBa_K1189031
). We then cultured bacteria in the treated LB that only carry resistance to chloramphenicol. Therefore, the bacteria are only able to survive if the our isolated protein retained its enzymatic abilities. We can show that the bacteria susceptible to ampicillin was able to grow in the presence of our purified construct protein (
BBa_K1189031
), which means that we are expressing and purifying functional protein which is degrading the ampicillin (Figures 1 and 3). Figure 3 shows the OD at 24 hour time point from culturing where Figure 1 shows OD change over time. Both graphs show an increase in OD for cultures pre-treated with our protein demonstrating our protein is functional. Figure 2. On the left crude lysate of beta-lactamase + His (
BBa_K1189007
) from different lysis protocols: a
mechanical
and with
sucrose
, respectively. On the right, western blot of
TALE A
-linker-beta-lactamase (
BBa_K1189031
) showing that we were able to express and purify our construct.
Figure 3. Absorbance values at 600nm after 24h. Amounts from 0.1µg to 20µg of TALE A-link-Beta-lactamase (
BBa_K1189031
) were sufficient to degrade the ampicillin in the media allowing bacteria susceptible to ampicillin to grow.
Figure 4. Absorbance values at 600nm in different time points. Amounts from 1.0µg to 10µg of TALE A-link-Beta-lactamase (
BBa_K1189031
) were sufficient to degrade the ampicillin in the media allowing bacteria susceptible to ampicillin to grow.
After verifying that
TALE A
-linker-beta-lactamase (
BBa_K1189031
) retained enzymatic activity and was able to degrade ampicillin, we performed a
colourimetric assay
using benzylpenicillin as our substrate. We were able to see a colour change from red to yellow. This is because there is phenol red, a pH indicator, added to the substrate solution. Beta-lactamase hydrolyzes benzylpenicillin to penicillinoic acid, which changes the pH of the solution from alkaline to acidic. This pH change causes the phenol red to change from red to yellow. Our negative controls, to which benzylpenicillin was not added, remained red. We can also see the colour change correlate to the amount of purified TALE A linked to beta-lactamase present in each sample (Figure 5). Figure 5. Benzylpenicillin assay. On the top, the wells only had
TALE A
-linker-beta-lactamase (
BBa_K1189031
). Benzylpenicillin was added and after a 10-minute incubation at room temperature, we were able to observe a colour output from red to yellow (bottom row) while the control wells remained red.





Additionally, we have demonstrated the same pH change of benzylpenicillin to penicillinoic acid by the TALE A β-lactamase fusion (BBa_K1189031) with bromothymol blue. The pH change causes the bromothymol blue to change in colour from blue to yellow as it gets more acidic (Figure 11 & 12). In the presence of TALE A β-lactamase fusion we see the colour change to yellow whereas the negatives not containing TALE A β-lactamase fusion remains blue (Figure 11). We did a kinetic analysis at 616 nm every 30 seconds and as the blue colour disappears, the absorbance at 616 nm decreases (Figure 12). Therefore, the lower the decrease the better the TALE A β-lactamase fusion reporter activity. Our kinetic activity shows that the biggest decrease is in our positive recombinant β-lactamase followed by the TALE A β-lactamase fusion at 10 micrograms. This decrease is lessened as we decrease the amount of TALE A β-lactamase fusion. We can also show that in our negatives with no TALE A β-lactamase fusion we do not have a decrease in absorbance. We have demonstrated the reporter activity both qualitatively (Figure 10 & 11) and quantitatively (Figure 12).

Figure 11.Change in pH catalyzed by TALE A linked to β-lactamase (BBa_K1189031) using benzylpenicillin. Bromothymol blue was used to keep track of this colour change. Absorbance readings were taken at 616 nm every 30 seconds. Different amounts of TALE A linked to β-lactamase (BBa_K1189031) were tested. Commercial β-lactamase was used as a positive control. Negative controls included were bovine serum albumin, β-lactamase without the substrate and the substrate by itself.
In order to demonstrate that we can successfully capture target DNA with two TALEs we did a capture TALE assay. TALE B was incubated with DNA containing target sites for TALE A and TALE B and blotted on nitrocellulose. After blocking and washing, TALE A β-lactamase fusion (BBa_K1189031) was added to the nitrocellulose strips. The strips were transferred into a 96 well plate to which a benzylpenicillin substrate solution with phenol red was added. If the TALE A β-lactamase fusion bound to the target site for TALE A then the solution will change colour from pink to clear. If TALE A β-lactamase fusion was not present, the solution will remain pink. We can show that the first four samples which have TALE B with DNA for TALE A and TALE B show a colour change indicating that we are successfully capturing the target DNA and reporting it. Furthermore we can also show that when we add non-specific DNA we do not see a colour change demonstrating that we can successfully capture only specific DNA and report its presence with an easy visual colourimetric output.
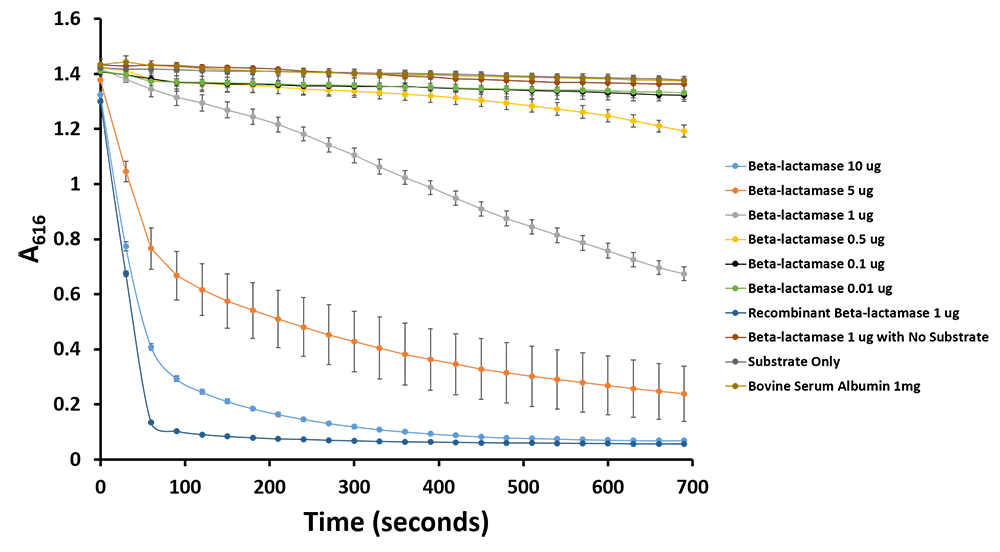
Figure 12.Change in pH catalyzed by TALE A linked to β-lactamase (BBa_K1189031) using benzylpenicillin. Bromothymol blue was used to keep track of this colour change. Absorbance readings were taken at 616 nm every 30 seconds. Different amounts of TALE A linked to β-lactamase (BBa_K1189031) were tested. Commercial β-lactamase was used as a positive control. Negative controls included were bovine serum albumin, β-lactamase without the substrate and the substrate by itself.

Figure 6: TALE capture assay was done with TALE B ( BBa_K1189001 )and TALE A B-lac fusion ( BBa_K1189031 ) and DNA ( BBa_K1189006 ) with both target sequences. If capture is successful, the B-lac is present in the well giving a colour change from pink to yellow when subjected to benzylpenicillin substrate solution within 20 minutes. The only wells that change colour are the first four wells which contain TALE B, specific DNA, and TALE A B-lac fusion and our positive control wells which are the controls for our fusion TALE A B-lac protein and our positive recombinant b-lac. All our other controls including our test using a non-specific sequence of DNA remained pink . This preliminary characterization data demonstrates that the TALEs are able to bind to DNA with specificity. Additionally it also shows that our system of capturing DNA with two detector TALEs and then subsequent reporting of the DNA’s presence works.
This assay shows that we can capture our target DNA with two detector TALEs with specificity . Additionally, we can report whether that DNA has been captured and is present in the sample, which is a very important concept for our sensor system.
To conclude, we have demonstrated that we can build, express, and purify this part. This biobrick has also been characterized to show both its ability to be a good reporter and its ability to be able to bind to DNA with specificity.
To further see if the TALE capture assay works (figure 17). In order to do this, we blotted TALEB fused to a K coil (BBa_K1189030) on a nitrocellulose strip. Subsequently the strips were blocked with 5% skimmed milk in TBST buffer. The strips were then washed and soaked in a solution containing [A] and [B] on the same plasmid(BBa_K1189006). The strips were then washed again with TALE binding buffer and soaked in a solution containing TALEA+β-lac (BBa_K1189031). Then a dot blot was done on the strips with β-lactamase primary antibody and an hrp conjugated secondary antibody. This assay demonstrated that the TALEs can capture DNA.

Figure 7. Figure on the left: For the dual TALE lanes TALEB fused to a K coil (BBa_K1189030) was blotted on a nitrocellulose strip. Subsequently the strips were blocked with 5% skimmed milk in TBST buffer. The strips were then washed and soaked in a solution containing [A] and [B] on the same plasmid(BBa_K1189006). The strips were then washed again with TALE binding buffer and soaked in a solution containing TALEA+β-lac (BBa_K1189031). Then a blot dot was done on the strips with β-lactamase primary antibody and an hrp conjugated secondary antibody. In negative 1 the strip was not soaked in the DNA solution. In Negative 2, the strip was not soaked in TALEA+β-lac solution. For positive controls, TALEA+βlac was blotted on the strip at the beginning instead of TALEB+K coil. The figure on the right shows the result of the densitometery we performed on the dot blot assay.
User Reviews
UNIQ9d34b99389a360f3-partinfo-00000003-QINU UNIQ9d34b99389a360f3-partinfo-00000004-QINU