Difference between revisions of "Part:BBa K1150034"
Lisaschmunk (Talk | contribs) |
|||
| (14 intermediate revisions by 6 users not shown) | |||
| Line 32: | Line 32: | ||
|} | |} | ||
| − | This device contains loci | + | This device contains loci coding for small RNAs that are able to target the catalytically-inactive, engineered [https://parts.igem.org/Part:BBa_K1150000 dCas9] protein to any desired locus adjacent to a so-called PAM sequence (NGG). The plasmid consists of a human H1 promoter driving the expression of the tracrRNA which is mediating the interaction between the dCas9 protein and the second RNA, the crRNA. This crRNA gives rise to the specific binding of dCas9 to DNA and is under control of the human U6 promoter. The 30 bp target site can be inserted by simply opening the plasmid with BbsI and ligating the desired sequence into the plasmid. The flanking direct repeats (DR) will mediate interaction with the tracrRNA. <br> |
| − | Multiple target sites can be combined on one plasmid by using the BioBrick standard and enables | + | Multiple target sites can be combined on one plasmid by using the BioBrick standard and enables targeting of multiple loci at once with a small amount of plasmids. <br> |
| + | The minimalistic pSB1C3 backbone guarantees strong expression of the RNAs, which are suspected to be the limiting factor of dCas9 interaction. <br> | ||
| + | This plasmid should be used in combination of dCas9-effector fusion proteins. They can be found in the registry or on [http://2013.igem.org/Team:Freiburg/Project/effector our homepage]. <br> | ||
| − | This year's Freiburg iGEM team 2013 | + | This year's Freiburg iGEM team 2013 furthermore provides an efficient tool for designing target sites applicable to the CRISPR/Cas system, the [http://2013.igem.org/Team:Freiburg/Project/crrna#design_tool crRNA design tool]. |
<br> | <br> | ||
===Assembly=== | ===Assembly=== | ||
| − | <p> On this plasmid two RNAs are encoded. The tracrRNA is mediating the interaction between dCas9 and the crRNA. The direct repeats (DRs) of the crRNA are complementary to regions of tracrRNA. The inserted locus in the crRNA is complementary to the locus | + | <p> On this plasmid two RNAs are encoded. The tracrRNA is mediating the interaction between dCas9 and the crRNA. The direct repeats (DRs) of the crRNA are complementary to regions of tracrRNA. The inserted locus in the crRNA is complementary to the locus dCas9 should target. On this plasmid a 10 bp long dummy-DNA is inserted, which has two BbsI cleavage sites. After digesting with BbsI the locus is opened and the DRs end with sticky ends, so new targets can be easily ligated. For the generation of target sites annealed oligos will work fine. <br> |
| − | Once single loci are inserted it is | + | Once single loci are inserted it is easy to combine several loci on one plasmid using the idempotent iGEM cloning. <br> |
| − | + | For a more detailed introduction to our system [http://2013.igem.org/Team:Freiburg/Project/crrna#rnaimer click here]. <br> | |
[[File:Multiple_targeting_Freiburg_2013_%281%29.png|800px|]] <br> | [[File:Multiple_targeting_Freiburg_2013_%281%29.png|800px|]] <br> | ||
| − | '''Figure 1 | + | '''Figure 1:''' Scheme of the RNAimer. It contains the two RNA encoding loci for tracrRNA and crRNA. Into the crRNA a desired target site can be inserted via ligation of annealed oligos. |
<br> <br> <br> | <br> <br> <br> | ||
=== Proof of functionality === | === Proof of functionality === | ||
| − | To test the functionality of this plasmid we used the | + | To test the functionality of this plasmid we used the [https://parts.igem.org/Part:BBa_K1150020 dCas9-VP16] construct in combination with various targets. We tested if it is possible to activate transcription of a secrected alkaline phosphatase (SEAP) reporter with this RNA plasmid. If functional, we expected an increase in SEAP activity in the supernatant. Cells were seeded in 24-well plates at a density of 65,000 cells. 24 hours later cells were transfected with a CMV<sub>min</sub>:SEAP and RNAimer plasmids containing different target sites. 48 hours post-transfection, SEAP was assayed as described in <html> <a id="link" href="http://2013.igem.org/Team:Freiburg/protocols#SEAP Assay"> our standard protocol</a></html>. |
[[File:Multiple_Targeting_Freiburg_2013_%285%29.png|600px|]] <br> | [[File:Multiple_Targeting_Freiburg_2013_%285%29.png|600px|]] <br> | ||
| − | '''Figure 2 | + | '''Figure 2:''' Activation via dCas9-VP16 in combination with the RNAimer plasmid. Experiment has been conducted as decribed above. It is clearly visible that activation depends on which target site is ligated into the RNAimer, resulting in different binding of the dCas9-VP16 on the plasmid. Each target was cotransfected on one RNAimer plasmid. On the fourth bar two plasmids were co-transfected. The observed effect exceeds the additive effect of two loci. <br> <br> |
| − | + | A main feature of the CRISPR/Cas9 system is the ability to target multiple loci at once. To test whether this is possible putting more than one target site on the same RNAimer-derived plasmid we tested activation of the SEAP promoter using two single loci plasmids and an engineered RNAimer-plasmid carrying two target sites, assembled via the BioBrick idempotent cloning standard.<br> | |
[[File:Multiple_Targeting_Freiburg_2013_%287%29.png|600px|]] <br> | [[File:Multiple_Targeting_Freiburg_2013_%287%29.png|600px|]] <br> | ||
| − | '''Figure 3 | + | '''Figure 3:''' Activation of SEAP expression with two targets either with two seperate plasmids or with both target sites on the same RNAimer plasmid. As shown, no difference in activation could be measured. Combining several targets on one plasmid does not seem to affect the effectivity of targeting. <br><br> |
| + | |||
| + | As we see in figure 3 there is no difference in activation of a SEAP reporter whether we transfect the target sites on seperate plasmids or on a combined RNAimer plasmid. This provides the great possibility to reduce plasmid numbers in transfections. Furthermore, this clearly demonstrates the functionality of our designed concept and the engineered RNAimer. <br><br> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<span class='h3bb'> | <span class='h3bb'> | ||
| + | |||
==Sequence and Features== | ==Sequence and Features== | ||
</span> | </span> | ||
<partinfo>BBa_K1150034 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1150034 SequenceAndFeatures</partinfo> | ||
| − | == | + | ==References== |
<small> | <small> | ||
| + | [1]Cong, L., Ran, F.A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P.D., Wu, X., Jiang, W., Marraffini, L.A., Zhang, F. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339 (6121), 819-23 <br> | ||
| + | </small> | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 02:26, 29 October 2013
dCas RNA plasmid: "uniCAS RNAimer"
| pH1:tracrRNA pU6:DR_dummy_DR | |
|---|---|
| Function | two small RNAs giving rise to
specific binding of dCas9 to DNA loci |
| Use in | Mammalian cells |
| RFC standard | RFC 25 |
| Backbone | pSB1C3 |
| Organism | Streptococcus pyogenes |
| Source | Feng Zhang, Addgene |
| Submitted by | [http://2013.igem.org/Team:Freiburg Freiburg 2013] |
This device contains loci coding for small RNAs that are able to target the catalytically-inactive, engineered dCas9 protein to any desired locus adjacent to a so-called PAM sequence (NGG). The plasmid consists of a human H1 promoter driving the expression of the tracrRNA which is mediating the interaction between the dCas9 protein and the second RNA, the crRNA. This crRNA gives rise to the specific binding of dCas9 to DNA and is under control of the human U6 promoter. The 30 bp target site can be inserted by simply opening the plasmid with BbsI and ligating the desired sequence into the plasmid. The flanking direct repeats (DR) will mediate interaction with the tracrRNA.
Multiple target sites can be combined on one plasmid by using the BioBrick standard and enables targeting of multiple loci at once with a small amount of plasmids.
The minimalistic pSB1C3 backbone guarantees strong expression of the RNAs, which are suspected to be the limiting factor of dCas9 interaction.
This plasmid should be used in combination of dCas9-effector fusion proteins. They can be found in the registry or on [http://2013.igem.org/Team:Freiburg/Project/effector our homepage].
This year's Freiburg iGEM team 2013 furthermore provides an efficient tool for designing target sites applicable to the CRISPR/Cas system, the [http://2013.igem.org/Team:Freiburg/Project/crrna#design_tool crRNA design tool].
Assembly
On this plasmid two RNAs are encoded. The tracrRNA is mediating the interaction between dCas9 and the crRNA. The direct repeats (DRs) of the crRNA are complementary to regions of tracrRNA. The inserted locus in the crRNA is complementary to the locus dCas9 should target. On this plasmid a 10 bp long dummy-DNA is inserted, which has two BbsI cleavage sites. After digesting with BbsI the locus is opened and the DRs end with sticky ends, so new targets can be easily ligated. For the generation of target sites annealed oligos will work fine.
Once single loci are inserted it is easy to combine several loci on one plasmid using the idempotent iGEM cloning.
For a more detailed introduction to our system [http://2013.igem.org/Team:Freiburg/Project/crrna#rnaimer click here].
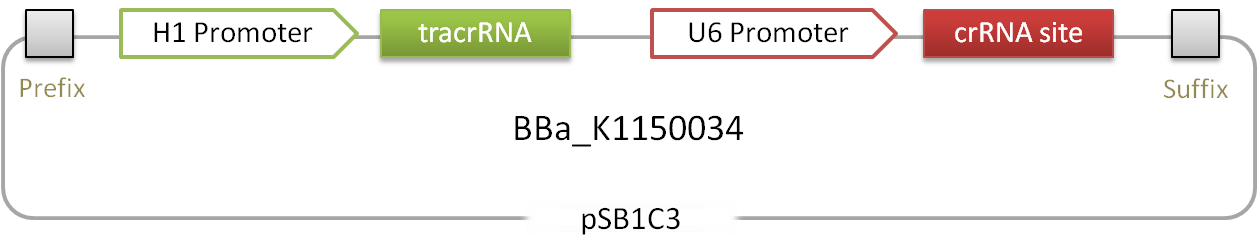
Figure 1: Scheme of the RNAimer. It contains the two RNA encoding loci for tracrRNA and crRNA. Into the crRNA a desired target site can be inserted via ligation of annealed oligos.
Proof of functionality
To test the functionality of this plasmid we used the dCas9-VP16 construct in combination with various targets. We tested if it is possible to activate transcription of a secrected alkaline phosphatase (SEAP) reporter with this RNA plasmid. If functional, we expected an increase in SEAP activity in the supernatant. Cells were seeded in 24-well plates at a density of 65,000 cells. 24 hours later cells were transfected with a CMVmin:SEAP and RNAimer plasmids containing different target sites. 48 hours post-transfection, SEAP was assayed as described in our standard protocol.
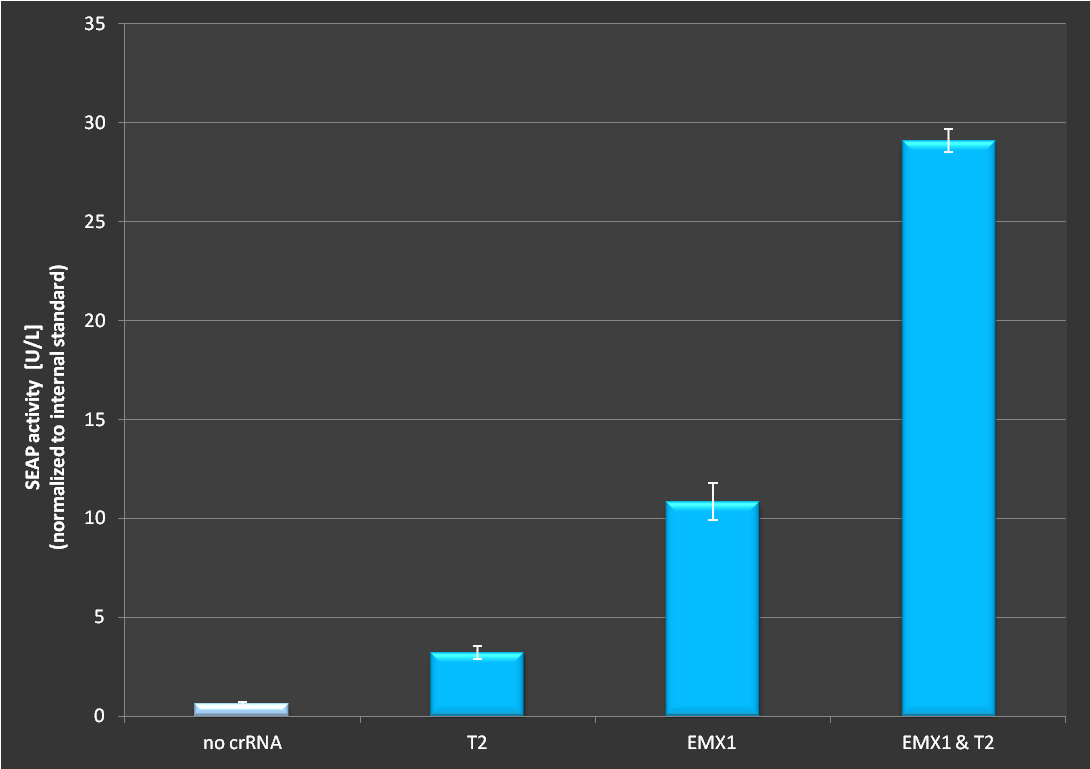
Figure 2: Activation via dCas9-VP16 in combination with the RNAimer plasmid. Experiment has been conducted as decribed above. It is clearly visible that activation depends on which target site is ligated into the RNAimer, resulting in different binding of the dCas9-VP16 on the plasmid. Each target was cotransfected on one RNAimer plasmid. On the fourth bar two plasmids were co-transfected. The observed effect exceeds the additive effect of two loci.
A main feature of the CRISPR/Cas9 system is the ability to target multiple loci at once. To test whether this is possible putting more than one target site on the same RNAimer-derived plasmid we tested activation of the SEAP promoter using two single loci plasmids and an engineered RNAimer-plasmid carrying two target sites, assembled via the BioBrick idempotent cloning standard.
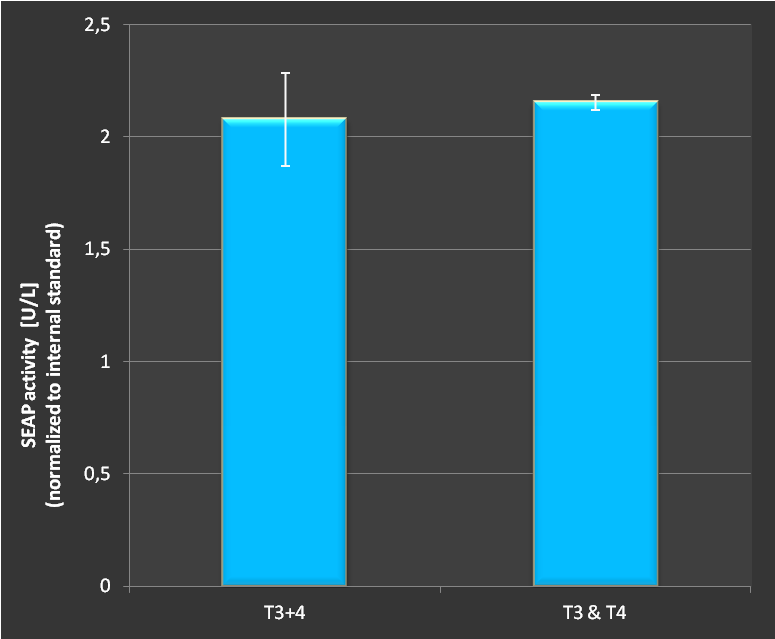
Figure 3: Activation of SEAP expression with two targets either with two seperate plasmids or with both target sites on the same RNAimer plasmid. As shown, no difference in activation could be measured. Combining several targets on one plasmid does not seem to affect the effectivity of targeting.
As we see in figure 3 there is no difference in activation of a SEAP reporter whether we transfect the target sites on seperate plasmids or on a combined RNAimer plasmid. This provides the great possibility to reduce plasmid numbers in transfections. Furthermore, this clearly demonstrates the functionality of our designed concept and the engineered RNAimer.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 224
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 216
References
[1]Cong, L., Ran, F.A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P.D., Wu, X., Jiang, W., Marraffini, L.A., Zhang, F. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339 (6121), 819-23
