Difference between revisions of "Part:BBa K1150003"
Lisaschmunk (Talk | contribs) |
Lisaschmunk (Talk | contribs) |
||
| (32 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1150003 short</partinfo> | <partinfo>BBa_K1150003 short</partinfo> | ||
| − | |||
{| style="color:black" cellpadding="6" cellspacing="1" border="2" align="right" | {| style="color:black" cellpadding="6" cellspacing="1" border="2" align="right" | ||
! colspan="2" style="background:#FFBF00;"|G9a | ! colspan="2" style="background:#FFBF00;"|G9a | ||
|- | |- | ||
|'''Function''' | |'''Function''' | ||
| − | |Histone | + | |Histone Methyltransferase |
|- | |- | ||
|'''Use in''' | |'''Use in''' | ||
| Line 18: | Line 17: | ||
|- | |- | ||
|'''Organism''' | |'''Organism''' | ||
| − | |Mus musculus | + | |<i>Mus musculus</i> |
|- | |- | ||
|'''Source''' | |'''Source''' | ||
| Line 27: | Line 26: | ||
|} | |} | ||
| − | |||
| − | <!-- Add more about the biology of this part here | + | This BioBrick is part of the [http://2013.igem.org/Team:Freiburg Freiburg 2013 uniCAS toolkit for gene regulation] and contains the histone methyltransferase (HMTase) G9a encoded by the murine <i>EHMT2</i> gene. G9a is able to induce heterochromatization by transferring methyl groups to lysine 9 of histone 3 (H3K9) [1]. This part only contains the catalytically active SET domain of G9a that is responsible for histone modification. By fusing an HMTase domain to the DNA binding dCas9 device, specific endogenous gene targeting is possible. In regions of open chromatin structures H3K9 methylation can therefore lead to inactivation of the concerned locus. In combination with a customized [https://parts.igem.org/Part:BBa_K1150034 RNAimer plasmid] different loci can be targeted for [http://2013.igem.org/Team:Freiburg/Project/effector#epigenetics transcriptional repression] of gene expression. |
| + | |||
| + | <!-- Add more about the biology of this part here --> | ||
| + | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | The murine <i>EHMT2 (Euchromatic histone-lysine N-methyltransferase 2)</i> gene encodes a mammalian lysine-preferring histone methyltransferase called G9a [1]. The complete G9a protein contains a SET (Su(var)3-9, Enhancer of Zeste, Trithorax) domain, which catalyzes transfer of methyl groups to H3K9 and an ANK domain which has a role in <i>de novo</i> DNA methylation [2]. The G9a BioBrick only comprises the G9a-SET domain. To provide the possibility of stable endogenous repression in our uniCAS toolkit we constructed the following four devices using the G9a BioBrick. | ||
| + | <ol> | ||
| + | <li> the [https://parts.igem.org/Part:BBa_K1150024 uniCAS Histone Modifier (CMV promoter)] combines a strong mammalian [https://parts.igem.org/Part:BBa_K747096 promoter] with [https://parts.igem.org/Part:BBa_K1150000 dCas9] and the G9a. When co-transfected with an [https://parts.igem.org/Part:BBa_K1150034 RNAimer plasmid] transcriptional repression of a target gene can be inferred by this device.</li> | ||
| + | <li> the [https://parts.igem.org/Part:BBa_K1150024 uniCAS Histone Modifier (SV40 promoter)] uses a medium strong SV40 promoter to optimize this device for means were a strong expression is suboptimal. </li> | ||
| + | <li> the [https://parts.igem.org/Part:BBa_K1150028 uniCAS Red Light Switch Part II - Histone Modifier] is a result of combining G9a with part of our red light receptor [https://parts.igem.org/Part:BBa_K1150004 Phytochrome B]. PhyB will dimerize with [https://parts.igem.org/Part:BBa_K1150005 PIF6] upon red light stimulus. PIF6 is fused to dCas9 in the [https://parts.igem.org/Part:BBa_K1150025 uniCAS Red Light Switch Part I - Stimulator]. Using these two devices G9a is brought into proximity of the DNA targeting dCas9 and should therefore be able to site specifically transfer methyl groups to histone H3 and to repress gene expression. | ||
| + | <li> the [https://parts.igem.org/Part:BBa_K1150032 uniCAS UV Light Switch Part II - Histone Modifier] combines G9a with [https://parts.igem.org/Part:BBa_K1150007 COP1] which can dimerize with [https://parts.igem.org/Part:BBa_K1150006 UVR8] on [https://parts.igem.org/Part:BBa_K1150029 uniCAS UV Light Switch Part I - Stimulator] upon UV-light stimulus [3]. </li> | ||
| + | </ol> | ||
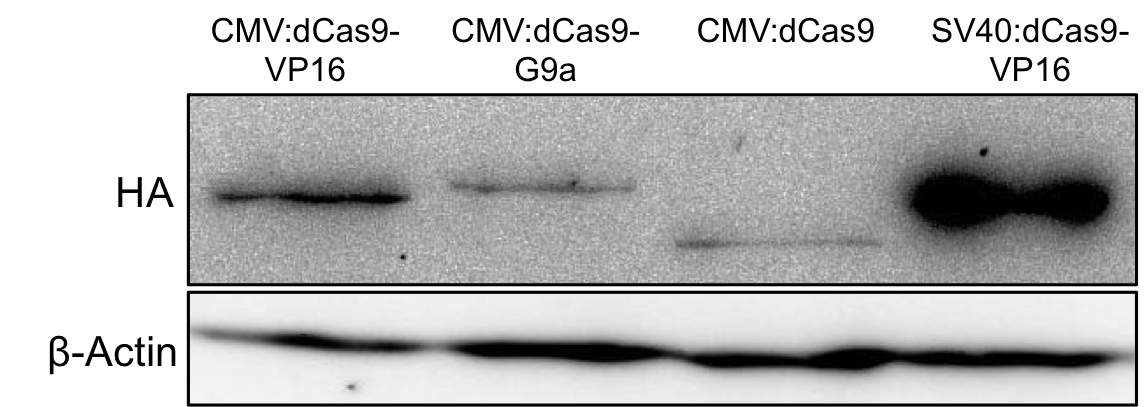
| − | < | + | ===Expression tests=== |
| − | <span class='h3bb'>Sequence and Features</span> | + | The expression of G9a fusion constructs was first determined via western blotting. Therefore mammalian HEK293T cells were transfected with the [https://parts.igem.org/Part:BBa_K1150024 uniCAS Histone Modifier (CMV promoter)] in 6-well plates (150,000 cells/well). 48h after transfection the cells were harvested and the complete cell lysate was blotted with anti-HA antibody against the N-terminally HA-tagged dCas9 proteins. The fullength dCas9-G9a protein could be detected at the expected size of approximately 185kDa. |
| + | |||
| + | [[File:UniBAssblotfertig2.1-team_Freiburgforregistry.PNG|700px|]] | ||
| + | |||
| + | '''Figure 1:''' HA-tag fused to different dCas9-fusion proteins encoded on the RFC25 pSB1C3 backbone. dCas9-G9a under a CMV promoter is expressed at the expected size (185 kDa). | ||
| + | |||
| + | |||
| + | ===Functional tests=== | ||
| + | Targeting of dCas9-G9a to an open, endogenous locus should lead to changes in the epigenetic chromatin state and repression of gene expression. To test this, the endogenous <b>VEGF-A locus </b> was chosen, as it is well characterized and open in HEK-293T cells. Additionally, VEGF production can easily be measured by ELISA analysis. <br> Cells were seeded into 24-well format at a density of 65,000 cells per well and were co-transfected with the desired crRNAs, based on the [https://parts.igem.org/Part:BBa_K1150034 RNAimer]. These plasmids are derivates of the RNAimer, the only difference was the insertion of the target sites. These were: [https://parts.igem.org/Part:BBa_K1150036 -8], [https://parts.igem.org/Part:BBa_K1150037 -573], [https://parts.igem.org/Part:BBa_K1150038 +434], [https://parts.igem.org/Part:BBa_K1150039 -475] and [https://parts.igem.org/Part:BBa_K1150043 -573/+434]. As an off-target control crRNA targeting [https://parts.igem.org/Part:BBa_K1150035 EMX] was used. Every site was targeted with [https://parts.igem.org/Part:BBa_K1150017 CMV:dCas9] wthout G9a, to differentiate the action of G9a from the CRISPRi effect. As an <b> internal standard </b> a constitutive SEAP reporter was used. 24 hours post transfection, VEGF levels and SEAP levels were measured and the ratio was calculated. Error bars represent standard deviations. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | [[File:Freiburg2013_G9a_test_neu.jpg|800px|]] <br> | ||
| + | '''Figure 2:''' dCas9-G9a targeting of endogenous VEGF-A locus in HEK-293T cells.<br>ELISA values, normalized to internal SEAP standard. For some loci clear repressive effects up to 50% were detectable. Controls did not display any off-target effects. Combining two target sites did not show stronger repression than targeting one site at a time.<br><br> | ||
| + | <br> | ||
| + | Different VEGF target sites led to different repression efficiencies and simultaneous targeting of two sites did not further enhance the observed effects. <br> In summary, the CMV:dCas9-G9a is an effective repressor of endogenous gene expression, by modulating the chromatin structure of the targeted locus. This offers application in fundamental research as well as in medical sciences. | ||
| + | |||
| + | |||
| + | <span class='h3bb'> | ||
| + | ==Sequence and Features== | ||
| + | </span> | ||
<partinfo>BBa_K1150003 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1150003 SequenceAndFeatures</partinfo> | ||
| + | <br> | ||
| + | <html><!--- Please copy this table containing parameters for BBa_ at the end of the parametrs section ahead of the references. ---><style type="text/css">table#AutoAnnotator {border:1px solid black; width:100%; border-collapse:collapse;} th#AutoAnnotatorHeader { border:1px solid black; width:100%; background-color: rgb(221, 221, 221);} td.AutoAnnotator1col { width:100%; border:1px solid black; } span.AutoAnnotatorSequence { font-family:'Courier New', Arial; } td.AutoAnnotatorSeqNum { text-align:right; width:2%; } td.AutoAnnotatorSeqSeq { width:98% } td.AutoAnnotatorSeqFeat1 { width:3% } td.AutoAnnotatorSeqFeat2a { width:27% } td.AutoAnnotatorSeqFeat2b { width:97% } td.AutoAnnotatorSeqFeat3 { width:70% } table.AutoAnnotatorNoBorder { border:0px; width:100%; border-collapse:collapse; } table.AutoAnnotatorWithBorder { border:1px solid black; width:100%; border-collapse:collapse; } td.AutoAnnotatorOuterAmino { border:0px solid black; width:20% } td.AutoAnnotatorInnerAmino { border:1px solid black; width:50% } td.AutoAnnotatorAminoCountingOuter { border:1px solid black; width:40%; } td.AutoAnnotatorBiochemParOuter { border:1px solid black; width:60%; } td.AutoAnnotatorAminoCountingInner1 { width: 7.5% } td.AutoAnnotatorAminoCountingInner2 { width:62.5% } td.AutoAnnotatorAminoCountingInner3 { width:30% } td.AutoAnnotatorBiochemParInner1 { width: 5% } td.AutoAnnotatorBiochemParInner2 { width:55% } td.AutoAnnotatorBiochemParInner3 { width:40% } td.AutoAnnotatorCodonUsage1 { width: 3% } td.AutoAnnotatorCodonUsage2 { width:14.2% } td.AutoAnnotatorCodonUsage3 { width:13.8% } td.AutoAnnotatorAlignment1 { width: 3% } td.AutoAnnotatorAlignment2 { width: 10% } td.AutoAnnotatorAlignment3 { width: 87% } td.AutoAnnotatorLocalizationOuter {border:1px solid black; width:40%} td.AutoAnnotatorGOOuter {border:1px solid black; width:60%} td.AutoAnnotatorLocalization1 { width: 7.5% } td.AutoAnnotatorLocalization2 { width: 22.5% } td.AutoAnnotatorLocalization3 { width: 70% } td.AutoAnnotatorGO1 { width: 5% } td.AutoAnnotatorGO2 { width: 35% } td.AutoAnnotatorGO3 { width: 60% } td.AutoAnnotatorPredFeat1 { width:3% } td.AutoAnnotatorPredFeat2a { width:27% } td.AutoAnnotatorPredFeat3 { width:70% } div.AutoAnnotator_trans { position:absolute; background:rgb(11,140,143); background-color:rgba(11,140,143, 0.8); height:5px; top:100px; } div.AutoAnnotator_sec_helix { position:absolute; background:rgb(102,0,102); background-color:rgba(102,0,102, 0.8); height:5px; top:110px; } div.AutoAnnotator_sec_strand { position:absolute; background:rgb(245,170,26); background-color:rgba(245,170,26, 1); height:5px; top:110px; } div.AutoAnnotator_acc_buried { position:absolute; background:rgb(89,168,15); background-color:rgba(89,168,15, 0.8); height:5px; top:120px; } div.AutoAnnotator_acc_exposed { position:absolute; background:rgb(0, 0, 255); background-color:rgba(0, 0, 255, 0.8); height:5px; top:120px; } div.AutoAnnotator_dis { position:absolute; text-align:center; font-family:Arial,Helvetica,sans-serif; background:rgb(255, 200, 0); background-color:rgba(255, 200, 0, 1); height:16px; width:16px; top:80px; border-radius:50%; } </style><div id='AutoAnnotator_container_1381279535746'><table id="AutoAnnotator"><tr><!-- Time stamp in ms since 1/1/1970 1381279535746 --><th id="AutoAnnotatorHeader" colspan="2">Protein data table for BioBrick <a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_<!------------------------Enter BioBrick number here------------------------>">BBa_<!------------------------Enter BioBrick number here------------------------></a> automatically created by the <a href="http://2013.igem.org/Team:TU-Munich/Results/AutoAnnotator">BioBrick-AutoAnnotator</a> version 1.0</th></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Nucleotide sequence</strong> in <strong>RFC 25</strong>, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein)<br><span class="AutoAnnotatorSequence"> <u><i>ATGGCCGGC</i>GAGAATGTA ... ATCAACACC<i>ACCGGT</i></u></span><br> <strong>ORF</strong> from nucleotide position -8 to 846 (excluding stop-codon)</td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Amino acid sequence:</strong> (RFC 25 scars in shown in bold, other sequence features underlined; both given below)<br><span class="AutoAnnotatorSequence"><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorSeqNum">1 <br>101 <br>201 </td><td class="AutoAnnotatorSeqSeq">MAGENVPIPCVNGVDGEPCPEDYKYISENCETSTMNIDRNITHLQHCTCVDDCSSSNCLCGQLSIRCWYDKDGRLLQEFNKIEPPLIFECNQACSCWRSC<br>KNRVVQSGIKVRLQLYRTAKMGWGVRALQTIPQGTFICEYVGELISDAEADVREDDSYLFDLDNKDGEVYCIDARYYGNISRFINHLCDPNIIPVRVFML<br>HQDLRFPRIAFFSSRDIRTGEELGFDYGDRFWDIKSKYFTCQCGSEKCKHSAEAIALEQSRLARLDPHPELLPDLSSLPPINTTG*</td></tr></table></span></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Sequence features:</strong> (with their position in the amino acid sequence, see the <a href="http://2013.igem.org/Team:TU-Munich/Results/Software/FeatureList">list of supported features</a>)<table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorSeqFeat1"></td><td class="AutoAnnotatorSeqFeat2b">None of the supported features appeared in the sequence</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Amino acid composition:</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Ala (A)</td><td class="AutoAnnotatorInnerAmino">12 (4.2%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Arg (R)</td><td class="AutoAnnotatorInnerAmino">19 (6.7%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Asn (N)</td><td class="AutoAnnotatorInnerAmino">14 (4.9%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Asp (D)</td><td class="AutoAnnotatorInnerAmino">23 (8.1%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Cys (C)</td><td class="AutoAnnotatorInnerAmino">19 (6.7%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Gln (Q)</td><td class="AutoAnnotatorInnerAmino">11 (3.9%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Glu (E)</td><td class="AutoAnnotatorInnerAmino">19 (6.7%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Gly (G)</td><td class="AutoAnnotatorInnerAmino">17 (6.0%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">His (H)</td><td class="AutoAnnotatorInnerAmino">6 (2.1%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Ile (I)</td><td class="AutoAnnotatorInnerAmino">21 (7.4%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Leu (L)</td><td class="AutoAnnotatorInnerAmino">23 (8.1%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Lys (K)</td><td class="AutoAnnotatorInnerAmino">11 (3.9%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Met (M)</td><td class="AutoAnnotatorInnerAmino">4 (1.4%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Phe (F)</td><td class="AutoAnnotatorInnerAmino">12 (4.2%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Pro (P)</td><td class="AutoAnnotatorInnerAmino">15 (5.3%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Ser (S)</td><td class="AutoAnnotatorInnerAmino">20 (7.0%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Thr (T)</td><td class="AutoAnnotatorInnerAmino">11 (3.9%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Trp (W)</td><td class="AutoAnnotatorInnerAmino">4 (1.4%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Tyr (Y)</td><td class="AutoAnnotatorInnerAmino">11 (3.9%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Val (V)</td><td class="AutoAnnotatorInnerAmino">13 (4.6%)</td></tr></table></td></tr></table></td></tr><tr><td class="AutoAnnotatorAminoCountingOuter"><strong>Amino acid counting</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Total number:</td><td class="AutoAnnotatorAminoCountingInner3">285</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Positively charged (Arg+Lys):</td><td class="AutoAnnotatorAminoCountingInner3">30 (10.5%)</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Negatively charged (Asp+Glu):</td><td class="AutoAnnotatorAminoCountingInner3">42 (14.7%)</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Aromatic (Phe+His+Try+Tyr):</td><td class="AutoAnnotatorAminoCountingInner3">33 (11.6%)</td></tr></table></td><td class="AutoAnnotatorBiochemParOuter"><strong>Biochemical parameters</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Atomic composition:</td><td class="AutoAnnotatorBiochemParInner3">C<sub>1420</sub>H<sub>2196</sub>N<sub>394</sub>O<sub>437</sub>S<sub>23</sub></td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Molecular mass [Da]:</td><td class="AutoAnnotatorBiochemParInner3">32516.8</td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Theoretical pI:</td><td class="AutoAnnotatorBiochemParInner3">5.00</td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Extinction coefficient at 280 nm [M<sup>-1</sup> cm<sup>-1</sup>]:</td><td class="AutoAnnotatorBiochemParInner3">38390 / 39578 (all Cys red/ox)</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges</strong> <input type='button' id='hydrophobicity_charge_button' onclick='show_or_hide_plot_1381279535746()' value='Show'><span id="hydrophobicity_charge_explanation"></span><div id="hydrophobicity_charge_container" style='display:none'><div id="hydrophobicity_charge_placeholder0" style="width:100%;height:150px"></div><div id="hydrophobicity_charge_placeholder1" style="width:100%;height:150px"></div></div></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Codon usage</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorCodonUsage1"></td><td class="AutoAnnotatorCodonUsage2">Organism:</td><td class="AutoAnnotatorCodonUsage3"><i>E. coli</i></td><td class="AutoAnnotatorCodonUsage3"><i>B. subtilis</i></td><td class="AutoAnnotatorCodonUsage3"><i>S. cerevisiae</i></td><td class="AutoAnnotatorCodonUsage3"><i>A. thaliana</i></td><td class="AutoAnnotatorCodonUsage3"><i>P. patens</i></td><td class="AutoAnnotatorCodonUsage3">Mammals</td></tr><tr><td class="AutoAnnotatorCodonUsage1"></td><td class="AutoAnnotatorCodonUsage2">Codon quality (<a href="http://en.wikipedia.org/wiki/Codon_Adaptation_Index">CAI</a>):</td><td class="AutoAnnotatorCodonUsage3">good (0.69)</td><td class="AutoAnnotatorCodonUsage3">good (0.67)</td><td class="AutoAnnotatorCodonUsage3">acceptable (0.52)</td><td class="AutoAnnotatorCodonUsage3">good (0.65)</td><td class="AutoAnnotatorCodonUsage3">excellent (0.86)</td><td class="AutoAnnotatorCodonUsage3">excellent (0.83)</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Alignments</strong> (obtained from <a href='http://predictprotein.org'>PredictProtein.org</a>)<br> There were no alignments for this protein in the data base. The BLAST search was initialized and should be ready in a few hours.</td></tr><tr><th id='AutoAnnotatorHeader' colspan="2"><strong>Predictions</strong> (obtained from <a href='http://predictprotein.org'>PredictProtein.org</a>)</th></tr><tr><td class="AutoAnnotator1col" colspan="2"> There were no predictions for this protein in the data base. The prediction was initialized and should be ready in a few hours.</td><tr><td class="AutoAnnotator1col" colspan="2"> The BioBrick-AutoAnnotator was created by <a href="http://2013.igem.org/Team:TU-Munich">TU-Munich 2013</a> iGEM team. For more information please see the <a href="http://2013.igem.org/Team:TU-Munich/Results/Software">documentation</a>.<br>If you have any questions, comments or suggestions, please leave us a <a href="http://2013.igem.org/Team:TU-Munich/Results/AutoAnnotator">comment</a>.</td></tr></table></div><br><!-- IMPORTANT: DON'T REMOVE THIS LINE, OTHERWISE NOT SUPPORTED FOR IE BEFORE 9 --><!--[if lte IE 8]><script language="javascript" type="text/javascript" src="excanvas.min.js"></script><![endif]--><script type='text/javascript' src='http://code.jquery.com/jquery-1.10.0.min.js'></script><script type='text/javascript' src='http://2013.igem.org/Team:TU-Munich/Flot.js?action=raw&ctype=text/js'></script><script>function show_or_hide_plot_1381279535746(){hydrophobicity_datapoints = [[2.5,-0.74],[3.5,-0.28],[4.5,-0.96],[5.5,0.02],[6.5,0.40],[7.5,1.60],[8.5,1.60],[9.5,1.22],[10.5,0.24],[11.5,1.40],[12.5,0.20],[13.5,-0.72],[14.5,-0.72],[15.5,-0.96],[16.5,-1.30],[17.5,-0.92],[18.5,-1.54],[19.5,-1.54],[20.5,-1.48],[21.5,-2.76],[22.5,-2.70],[23.5,-1.10],[24.5,-0.56],[25.5,-1.00],[26.5,-0.92],[27.5,-0.16],[28.5,-1.76],[29.5,-1.74],[30.5,-1.20],[31.5,-0.64],[32.5,-0.76],[33.5,-0.76],[34.5,0.28],[35.5,-0.26],[36.5,-1.02],[37.5,-2.10],[38.5,-0.50],[39.5,-1.54],[40.5,-1.48],[41.5,0.18],[42.5,0.18],[43.5,-1.36],[44.5,-0.72],[45.5,-0.22],[46.5,-0.48],[47.5,1.06],[48.5,1.00],[49.5,-0.20],[50.5,0.44],[51.5,-0.22],[52.5,-1.22],[53.5,-0.68],[54.5,-0.68],[55.5,-0.68],[56.5,0.24],[57.5,0.90],[58.5,0.98],[59.5,0.98],[60.5,1.24],[61.5,0.32],[62.5,0.72],[63.5,-0.10],[64.5,1.10],[65.5,0.16],[66.5,0.06],[67.5,-1.54],[68.5,-1.42],[69.5,-2.62],[70.5,-2.52],[71.5,-3.16],[72.5,-1.70],[73.5,-0.16],[74.5,-0.16],[75.5,-0.78],[76.5,0.68],[77.5,-0.78],[78.5,-2.32],[79.5,-0.72],[80.5,-0.72],[81.5,-1.60],[82.5,-1.22],[83.5,0.32],[84.5,0.32],[85.5,1.58],[86.5,1.20],[87.5,2.02],[88.5,0.56],[89.5,-1.04],[90.5,-1.24],[91.5,-0.04],[92.5,-0.70],[93.5,0.50],[94.5,1.02],[95.5,-0.24],[96.5,-0.90],[97.5,-0.24],[98.5,-1.52],[99.5,-2.04],[100.5,-2.04],[101.5,-1.04],[102.5,-0.70],[103.5,-0.62],[104.5,-0.08],[105.5,0.74],[106.5,0.80],[107.5,-0.82],[108.5,0.72],[109.5,-0.02],[110.5,0.82],[111.5,-0.78],[112.5,0.76],[113.5,-0.34],[114.5,-0.34],[115.5,-1.24],[116.5,-0.18],[117.5,-1.72],[118.5,-1.08],[119.5,-0.26],[120.5,-0.30],[121.5,-0.74],[122.5,0.88],[123.5,-0.40],[124.5,0.04],[125.5,0.98],[126.5,0.36],[127.5,-0.62],[128.5,1.18],[129.5,0.50],[130.5,-0.96],[131.5,-0.34],[132.5,-0.34],[133.5,-0.68],[134.5,0.54],[135.5,1.74],[136.5,1.12],[137.5,1.00],[138.5,1.28],[139.5,0.30],[140.5,-0.90],[141.5,0.56],[142.5,1.72],[143.5,0.72],[144.5,0.10],[145.5,1.16],[146.5,-0.30],[147.5,-0.84],[148.5,-1.38],[149.5,0.16],[150.5,-1.10],[151.5,-1.10],[152.5,-2.16],[153.5,-2.16],[154.5,-3.16],[155.5,-2.52],[156.5,-1.06],[157.5,0.20],[158.5,0.20],[159.5,1.12],[160.5,0.68],[161.5,-0.78],[162.5,-2.12],[163.5,-2.12],[164.5,-2.96],[165.5,-2.96],[166.5,-1.42],[167.5,-0.90],[168.5,0.30],[169.5,1.28],[170.5,1.28],[171.5,0.80],[172.5,0.16],[173.5,-0.60],[174.5,-1.76],[175.5,-1.14],[176.5,-2.20],[177.5,-0.40],[178.5,-0.30],[179.5,-0.94],[180.5,-0.30],[181.5,1.30],[182.5,-0.30],[183.5,-0.78],[184.5,0.88],[185.5,0.82],[186.5,-0.78],[187.5,-0.40],[188.5,-0.46],[189.5,-0.32],[190.5,0.08],[191.5,0.46],[192.5,1.62],[193.5,1.42],[194.5,1.36],[195.5,1.02],[196.5,1.72],[197.5,1.64],[198.5,1.90],[199.5,0.36],[200.5,-0.90],[201.5,-0.52],[202.5,-2.18],[203.5,-0.98],[204.5,-0.60],[205.5,-0.80],[206.5,-0.66],[207.5,0.60],[208.5,0.60],[209.5,1.48],[210.5,2.22],[211.5,1.16],[212.5,-0.10],[213.5,-1.36],[214.5,-1.02],[215.5,-1.76],[216.5,-1.74],[217.5,-0.92],[218.5,-0.92],[219.5,-2.52],[220.5,-0.86],[221.5,-0.80],[222.5,-0.16],[223.5,-0.16],[224.5,0.28],[225.5,-0.56],[226.5,-1.18],[227.5,-2.64],[228.5,-1.38],[229.5,-1.30],[230.5,-1.92],[231.5,-0.32],[232.5,-0.20],[233.5,-0.92],[234.5,-1.52],[235.5,-1.08],[236.5,-1.42],[237.5,-0.78],[238.5,-0.12],[239.5,-0.04],[240.5,0.72],[241.5,0.08],[242.5,0.06],[243.5,-1.14],[244.5,-1.22],[245.5,-1.22],[246.5,-1.92],[247.5,-2.40],[248.5,-1.86],[249.5,-0.72],[250.5,-1.92],[251.5,-0.78],[252.5,0.76],[253.5,1.28],[254.5,1.68],[255.5,1.68],[256.5,0.62],[257.5,-0.44],[258.5,-1.70],[259.5,-1.70],[260.5,-0.64],[261.5,-0.84],[262.5,0.08],[263.5,0.28],[264.5,-0.80],[265.5,-1.80],[266.5,-1.22],[267.5,-2.68],[268.5,-1.22],[269.5,-0.14],[270.5,0.18],[271.5,-0.20],[272.5,1.26],[273.5,0.34],[274.5,-0.58],[275.5,0.50],[276.5,0.88],[277.5,-0.20],[278.5,0.86],[279.5,0.32],[280.5,-0.58],[281.5,-0.40],[282.5,-0.16]];charge_datapoints = [[2.5,-0.20],[3.5,-0.20],[4.5,-0.20],[5.5,-0.20],[6.5,0.00],[7.5,0.00],[8.5,0.00],[9.5,0.00],[10.5,0.00],[11.5,0.00],[12.5,-0.20],[13.5,-0.20],[14.5,-0.40],[15.5,-0.40],[16.5,-0.40],[17.5,-0.20],[18.5,-0.40],[19.5,-0.40],[20.5,-0.40],[21.5,-0.20],[22.5,-0.20],[23.5,0.00],[24.5,0.20],[25.5,0.00],[26.5,-0.20],[27.5,-0.20],[28.5,-0.40],[29.5,-0.40],[30.5,-0.20],[31.5,-0.20],[32.5,-0.20],[33.5,0.00],[34.5,0.00],[35.5,-0.20],[36.5,0.00],[37.5,0.00],[38.5,0.00],[39.5,0.00],[40.5,0.30],[41.5,0.10],[42.5,0.10],[43.5,0.20],[44.5,0.20],[45.5,0.10],[46.5,0.10],[47.5,0.10],[48.5,-0.20],[49.5,-0.40],[50.5,-0.40],[51.5,-0.40],[52.5,-0.40],[53.5,-0.20],[54.5,0.00],[55.5,0.00],[56.5,0.00],[57.5,0.00],[58.5,0.00],[59.5,0.00],[60.5,0.00],[61.5,0.00],[62.5,0.00],[63.5,0.20],[64.5,0.20],[65.5,0.20],[66.5,0.20],[67.5,0.00],[68.5,0.00],[69.5,-0.20],[70.5,-0.20],[71.5,0.00],[72.5,0.20],[73.5,0.00],[74.5,0.20],[75.5,0.00],[76.5,-0.20],[77.5,-0.20],[78.5,0.00],[79.5,0.00],[80.5,0.00],[81.5,0.00],[82.5,0.00],[83.5,-0.20],[84.5,-0.20],[85.5,0.00],[86.5,-0.20],[87.5,-0.20],[88.5,-0.20],[89.5,-0.20],[90.5,-0.20],[91.5,0.00],[92.5,0.00],[93.5,0.00],[94.5,0.00],[95.5,0.20],[96.5,0.20],[97.5,0.20],[98.5,0.40],[99.5,0.40],[100.5,0.40],[101.5,0.40],[102.5,0.40],[103.5,0.20],[104.5,0.20],[105.5,0.00],[106.5,0.00],[107.5,0.20],[108.5,0.20],[109.5,0.40],[110.5,0.40],[111.5,0.40],[112.5,0.20],[113.5,0.20],[114.5,0.20],[115.5,0.20],[116.5,0.20],[117.5,0.40],[118.5,0.40],[119.5,0.20],[120.5,0.20],[121.5,0.20],[122.5,0.00],[123.5,0.20],[124.5,0.20],[125.5,0.20],[126.5,0.20],[127.5,0.20],[128.5,0.00],[129.5,0.00],[130.5,0.00],[131.5,0.00],[132.5,0.00],[133.5,0.00],[134.5,0.00],[135.5,0.00],[136.5,-0.20],[137.5,-0.20],[138.5,-0.20],[139.5,-0.20],[140.5,-0.40],[141.5,-0.20],[142.5,-0.20],[143.5,-0.20],[144.5,-0.40],[145.5,-0.20],[146.5,-0.40],[147.5,-0.40],[148.5,-0.60],[149.5,-0.40],[150.5,-0.20],[151.5,-0.20],[152.5,-0.40],[153.5,-0.40],[154.5,-0.40],[155.5,-0.60],[156.5,-0.40],[157.5,-0.20],[158.5,-0.20],[159.5,-0.20],[160.5,-0.40],[161.5,-0.40],[162.5,-0.20],[163.5,-0.20],[164.5,-0.20],[165.5,-0.20],[166.5,-0.20],[167.5,-0.40],[168.5,-0.20],[169.5,-0.20],[170.5,-0.20],[171.5,-0.20],[172.5,-0.00],[173.5,-0.00],[174.5,-0.00],[175.5,0.20],[176.5,0.20],[177.5,-0.00],[178.5,-0.00],[179.5,0.20],[180.5,0.20],[181.5,0.20],[182.5,0.20],[183.5,0.30],[184.5,0.10],[185.5,0.10],[186.5,-0.10],[187.5,-0.10],[188.5,-0.20],[189.5,-0.20],[190.5,-0.20],[191.5,-0.00],[192.5,-0.00],[193.5,0.20],[194.5,0.20],[195.5,0.20],[196.5,0.20],[197.5,0.20],[198.5,0.10],[199.5,0.10],[200.5,-0.10],[201.5,-0.10],[202.5,0.10],[203.5,-0.00],[204.5,-0.00],[205.5,0.40],[206.5,0.40],[207.5,0.20],[208.5,0.20],[209.5,0.20],[210.5,-0.00],[211.5,-0.00],[212.5,0.20],[213.5,-0.00],[214.5,-0.00],[215.5,0.20],[216.5,0.20],[217.5,-0.00],[218.5,-0.00],[219.5,-0.20],[220.5,-0.40],[221.5,-0.40],[222.5,-0.40],[223.5,-0.40],[224.5,-0.20],[225.5,-0.20],[226.5,-0.40],[227.5,-0.20],[228.5,-0.00],[229.5,-0.00],[230.5,-0.20],[231.5,-0.00],[232.5,-0.00],[233.5,-0.00],[234.5,0.20],[235.5,0.40],[236.5,0.40],[237.5,0.20],[238.5,0.20],[239.5,-0.00],[240.5,-0.00],[241.5,-0.00],[242.5,-0.00],[243.5,-0.20],[244.5,-0.00],[245.5,-0.00],[246.5,0.20],[247.5,0.30],[248.5,0.50],[249.5,0.30],[250.5,0.10],[251.5,-0.10],[252.5,-0.20],[253.5,-0.20],[254.5,-0.20],[255.5,-0.20],[256.5,-0.20],[257.5,-0.20],[258.5,-0.00],[259.5,-0.00],[260.5,0.20],[261.5,0.40],[262.5,0.40],[263.5,-0.00],[264.5,-0.00],[265.5,0.10],[266.5,-0.10],[267.5,-0.30],[268.5,-0.10],[269.5,-0.10],[270.5,-0.20],[271.5,-0.40],[272.5,-0.20],[273.5,-0.20],[274.5,-0.20],[275.5,-0.20],[276.5,-0.00],[277.5,-0.00],[278.5,-0.00],[279.5,-0.00],[280.5,-0.00],[281.5,-0.00],[282.5,-0.00]];dis_datapoints = undefined;trans_datapoints = undefined;sec_helix_datapoints = undefined;sec_strand_datapoints = undefined;acc_exposed_datapoints = undefined;acc_buried_datapoints = undefined;flot_plot_options = []; flot_plot_options[0] = {grid: {borderWidth: {top: 0,right: 0,bottom: 0,left: 0}},legend: {show: false},xaxes: [{show: true,min: 0,max: 200,ticks: [[0.5, '1'], [24.5, '25'], [49.5, '50'], [74.5, '75'], [99.5, '100'], [124.5, '125'], [149.5, '150'], [174.5, '175'], [199.5, '200']],tickLength: -5}],yaxes: [{show: true,ticks: [[0, '0'], [4.5,'hydro-<br>phobic '], [-4.5,'hydro-<br>philic ']],min: -4.5,max: +4.5,font: {size: 12,lineHeight: 14,style: 'italic',weight: 'bold',family: 'sans-serif',variant: 'small-caps',color: 'rgba(100,149,237,1)'}},{show: true,ticks: [[0, ''], [1,'positive<br> charge'], [-1,'negative<br> charge']],position: 'right',min: -1,max: 1,font: {size: 12,lineHeight: 14,style: 'italic',weight: 'bold',family: 'sans-serif',variant: 'small-caps',color: 'rgba(255,99,71,1)'}}]};number_of_plots = 2;for ( plot_num = 1 ; plot_num < number_of_plots ; plot_num ++){flot_plot_options[plot_num] = $.extend(true, {} ,flot_plot_options[0]);flot_plot_options[plot_num].xaxes = [{min: plot_num*200,max: (plot_num + 1)*200,ticks: [ [plot_num*200 + 0.5, (plot_num*200 + 1).toString()], [plot_num*200 + 24.5, (plot_num*200 + 25).toString()], [plot_num*200 + 49.5, (plot_num*200 + 50).toString()], [plot_num*200 + 74.5, (plot_num*200 + 75).toString()], [plot_num*200 + 99.5, (plot_num*200 + 100).toString()], [plot_num*200 + 124.5, (plot_num*200 + 125).toString()], [plot_num*200 + 149.5, (plot_num*200 + 150).toString()], [plot_num*200 + 174.5, (plot_num*200 + 175).toString()], [plot_num*200 + 199.5, (plot_num*200 + 200).toString()] ],tickLength: -5}];};try {if( $('#AutoAnnotator_container_1381279535746 #hydrophobicity_charge_button').val() =='Show' ){$('#AutoAnnotator_container_1381279535746 #hydrophobicity_charge_container').css('display','block');$('#AutoAnnotator_container_1381279535746 #hydrophobicity_charge_button').val('Hide');var description_html = '<div id=\'AutoAnnotator_plot_selectors\'>';description_html = description_html + '<br> <input type=\'checkbox\' id=\'hydrophobicity_checkbox\' checked=\'checked\'> Moving average over 5 amino acids for hydrophobicity (<img src=\'https://static.igem.org/mediawiki/2013/e/e9/TUM13_hydrophobicity_icon.png\' alt=\'blue graph\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'charge_checkbox\' checked=\'checked\'> Moving average over 5 amino acids for charge (<img src=\'https://static.igem.org/mediawiki/2013/3/3e/TUM13_charge_icon.png\' alt=\'red graph\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'dis_checkbox\' checked=\'checked\'> Predicted disulfid bridges (<img src=\'https://static.igem.org/mediawiki/2013/2/28/TUM13_dis_icon.png\' alt=\'yellow circle\' height=\'10\'></img>) with the number of the bridge in the center';description_html = description_html + '<br> <input type=\'checkbox\' id=\'trans_checkbox\' checked=\'checked\'> Predicted transmembrane helices (<img src=\'https://static.igem.org/mediawiki/2013/7/78/TUM13_trans_icon.png\' alt=\'turquois bars\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'sec_checkbox\' checked=\'checked\'> Predicted secondary structure: Helices (<img src=\'https://static.igem.org/mediawiki/2013/b/bf/TUM13_helix_icon.png\' alt=\'violet bars\' height=\'10\'></img>) and beta-strands (<img src=\'https://static.igem.org/mediawiki/2013/b/bf/TUM13_strand_icon.png\' alt=\'yellow bars\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'acc_checkbox\' checked=\'checked\'> Predicted solvent accessability: Exposed (<img src=\'https://static.igem.org/mediawiki/2013/1/16/TUM13_exposed_icon.png\' alt=\'blue bars\' height=\'10\'></img>) and buried (<img src=\'https://static.igem.org/mediawiki/2013/0/0b/TUM13_buried_icon.png\' alt=\'green bars\' height=\'10\'></img>) residues';description_html = description_html + '<br></div>';$('#AutoAnnotator_container_1381279535746 #hydrophobicity_charge_explanation').html(description_html);plot_according_to_selectors_1381279535746();$('#AutoAnnotator_container_1381279535746 #AutoAnnotator_plot_selectors').find('input').click(plot_according_to_selectors_1381279535746);}else{$('#AutoAnnotator_container_1381279535746 #hydrophobicity_charge_container').css('display','none');$('#AutoAnnotator_container_1381279535746 #hydrophobicity_charge_button').val('Show');$('#AutoAnnotator_container_1381279535746 #hydrophobicity_charge_explanation').html('');}}catch(err){txt='There was an error with the button controlling the visibility of the plot.\n';txt=txt+'The originating error is:\n' + err + '\n\n';alert(txt);}};function plot_according_to_selectors_1381279535746(){try{var plot_datasets = [[],[]];if($('#AutoAnnotator_container_1381279535746 #hydrophobicity_checkbox').prop('checked') == true){plot_datasets[0] = { color: 'rgba(100,149,237,1)',data: hydrophobicity_datapoints,label: 'Hydrophobicity',lines: { show: true, fill: true, fillColor: 'rgba(100,149,237,0.1)' },yaxis: 1};}if($('#AutoAnnotator_container_1381279535746 #charge_checkbox').prop('checked') == true){plot_datasets[1] = {color: 'rgba(255,99,71,1)',data: charge_datapoints,label: 'Charge',lines: { show: true, fill: true, fillColor: 'rgba(255,99,71,0.1)' },yaxis: 2};}for (plot_num = 0 ; plot_num < number_of_plots ; plot_num ++){$.plot('#AutoAnnotator_container_1381279535746 #hydrophobicity_charge_placeholder'+ plot_num.toString(), plot_datasets, flot_plot_options[plot_num] );}var screen_width = $('canvas.flot-base').width(); var pos_of_first_tick = 46;var pos_of_last_tick = screen_width - 51;var tick_diff = (screen_width - 97)/199;if($('#AutoAnnotator_container_1381279535746 #dis_checkbox').prop('checked') == true){for ( j = 0 ; j < dis_datapoints.length ; j++ ){$('#AutoAnnotator_container_1381279535746 #hydrophobicity_charge_placeholder' + Math.floor((dis_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_dis\' style=\'left:' + ((pos_of_first_tick - 8 + (dis_datapoints[j][0] - 1)*tick_diff - Math.floor((dis_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px;\'><b>' + (j+1) + '</b></div>');$('#AutoAnnotator_container_1381279535746 #hydrophobicity_charge_placeholder' + Math.floor((dis_datapoints[j][1] - 1)/200) ).append('<div class=\'AutoAnnotator_dis\' style=\'left:' + ((pos_of_first_tick - 8 + (dis_datapoints[j][1] - 1)*tick_diff - Math.floor((dis_datapoints[j][1] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px;\'><b>' + (j+1) + '</b></div>');}}if($('#AutoAnnotator_container_1381279535746 #trans_checkbox').prop('checked') == true){for ( j = 0 ; j < trans_datapoints.length ; j++ ){$('#AutoAnnotator_container_1381279535746 #hydrophobicity_charge_placeholder' + Math.floor((trans_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_trans\' style=\'width:' + (((trans_datapoints[j][1] - trans_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px ;left:' + ((pos_of_first_tick + (trans_datapoints[j][0] - 1.5)*tick_diff - Math.floor((trans_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}}if($('#AutoAnnotator_container_1381279535746 #sec_checkbox').prop('checked') == true){for ( j = 0 ; j < sec_helix_datapoints.length ; j++ ){$('#AutoAnnotator_container_1381279535746 #hydrophobicity_charge_placeholder' + Math.floor((sec_helix_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_sec_helix\' style=\'width:' + (((sec_helix_datapoints[j][1] - sec_helix_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (sec_helix_datapoints[j][0] - 1.5)*tick_diff - Math.floor((sec_helix_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}for ( j = 0 ; j < sec_strand_datapoints.length ; j++ ){$('#AutoAnnotator_container_1381279535746 #hydrophobicity_charge_placeholder' + Math.floor((sec_strand_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_sec_strand\' style=\'width:' + (((sec_strand_datapoints[j][1] - sec_strand_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (sec_strand_datapoints[j][0] - 1.5)*tick_diff - Math.floor((sec_strand_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}}if($('#AutoAnnotator_container_1381279535746 #acc_checkbox').prop('checked') == true){for ( j = 0 ; j < acc_buried_datapoints.length ; j++ ){$('#AutoAnnotator_container_1381279535746 #hydrophobicity_charge_placeholder' + Math.floor((acc_buried_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_acc_buried\' style=\'width:' + (((acc_buried_datapoints[j][1] - acc_buried_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (acc_buried_datapoints[j][0] - 1.5)*tick_diff - Math.floor((acc_buried_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}for ( j = 0 ; j < acc_exposed_datapoints.length ; j++ ){$('#AutoAnnotator_container_1381279535746 #hydrophobicity_charge_placeholder' + Math.floor((acc_exposed_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_acc_exposed\' style=\'width:' + (((acc_exposed_datapoints[j][1] - acc_exposed_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (acc_exposed_datapoints[j][0] - 1.5)*tick_diff - Math.floor((acc_exposed_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}}}catch(err){txt='There was an error while drawing the selected elements for the plot.\n';txt=txt+'The originating error is:\n' + err + '\n\n';throw(txt);}}</script></html> | ||
| + | |||
| + | ==References== | ||
| + | <small> | ||
| + | <!-- | ||
| + | [1]Wolffe, A., et al. (1999). Epigenetics: Regulation Through Repression. Science 286169, 481. <br> | ||
| + | [2]Snowden, A., et al. (2002). Gene-Specific Targeting of H3K9 Methylation Is Sufficient for Initiating Repression In Vivo. Current Biology 12, 2159-2166. <br> | ||
| + | [3] Lee, D., et al. (2006). Histone 3 Lysine 9 Methyltransferase G9a Is a Transcriptional Coactivator for Nuclear Receptors. Journal of Biological Chemistry 281, 8476-8485. <br>--> | ||
| + | [1] Tachibana, M., et al. (2001). SET Domain-containing Protein, G9a, Is a Novel Lysine-preferring Mammalian Histone Methyltransferase with Hyperactivity and Specific Selectivity to Lysines 9 and 27 of Histone H3. The Journal of Biological Chemistry, 276, 25309-25317.<br> | ||
| + | [2] Epsztejn-Litman, S., et al. (2008). De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 15(11): 1176–1183. <br> | ||
| + | [3] Rizzini L., Favory J.-J., Cloix C., Faggionato D., O'Hara A., Kaiserli E., Baumeister R., Schäfer E., Nagy F., Jenkins G. I., Ulm R. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332 <br> | ||
| + | </small> | ||
| + | |||
| + | <!-- Design: This part was PCR amplified from a plasmid containing the murine G9a set-domain (A. Jeltsch, Stuttgart). Primers with overhangs containing the RFC_25 prefix and suffix (link?) were used to insert iGEM restriction sites. Two point mutations were introduced to eliminate illegal PstI restriction sites within the coding region of G9a (link sequence annotation). | ||
| + | |||
| + | |||
Latest revision as of 02:10, 29 October 2013
G9a
| G9a | |
|---|---|
| Function | Histone Methyltransferase |
| Use in | Mammalian cells |
| RFC standard | RFC 25 |
| Backbone | pSB1C3 |
| Organism | Mus musculus |
| Source | Albert Jeltsch, Stuttgart |
| Submitted by | [http://2013.igem.org/Team:Freiburg Freiburg 2013] |
This BioBrick is part of the [http://2013.igem.org/Team:Freiburg Freiburg 2013 uniCAS toolkit for gene regulation] and contains the histone methyltransferase (HMTase) G9a encoded by the murine EHMT2 gene. G9a is able to induce heterochromatization by transferring methyl groups to lysine 9 of histone 3 (H3K9) [1]. This part only contains the catalytically active SET domain of G9a that is responsible for histone modification. By fusing an HMTase domain to the DNA binding dCas9 device, specific endogenous gene targeting is possible. In regions of open chromatin structures H3K9 methylation can therefore lead to inactivation of the concerned locus. In combination with a customized RNAimer plasmid different loci can be targeted for [http://2013.igem.org/Team:Freiburg/Project/effector#epigenetics transcriptional repression] of gene expression.
Usage and Biology
The murine EHMT2 (Euchromatic histone-lysine N-methyltransferase 2) gene encodes a mammalian lysine-preferring histone methyltransferase called G9a [1]. The complete G9a protein contains a SET (Su(var)3-9, Enhancer of Zeste, Trithorax) domain, which catalyzes transfer of methyl groups to H3K9 and an ANK domain which has a role in de novo DNA methylation [2]. The G9a BioBrick only comprises the G9a-SET domain. To provide the possibility of stable endogenous repression in our uniCAS toolkit we constructed the following four devices using the G9a BioBrick.
- the uniCAS Histone Modifier (CMV promoter) combines a strong mammalian promoter with dCas9 and the G9a. When co-transfected with an RNAimer plasmid transcriptional repression of a target gene can be inferred by this device.
- the uniCAS Histone Modifier (SV40 promoter) uses a medium strong SV40 promoter to optimize this device for means were a strong expression is suboptimal.
- the uniCAS Red Light Switch Part II - Histone Modifier is a result of combining G9a with part of our red light receptor Phytochrome B. PhyB will dimerize with PIF6 upon red light stimulus. PIF6 is fused to dCas9 in the uniCAS Red Light Switch Part I - Stimulator. Using these two devices G9a is brought into proximity of the DNA targeting dCas9 and should therefore be able to site specifically transfer methyl groups to histone H3 and to repress gene expression.
- the uniCAS UV Light Switch Part II - Histone Modifier combines G9a with COP1 which can dimerize with UVR8 on uniCAS UV Light Switch Part I - Stimulator upon UV-light stimulus [3].
Expression tests
The expression of G9a fusion constructs was first determined via western blotting. Therefore mammalian HEK293T cells were transfected with the uniCAS Histone Modifier (CMV promoter) in 6-well plates (150,000 cells/well). 48h after transfection the cells were harvested and the complete cell lysate was blotted with anti-HA antibody against the N-terminally HA-tagged dCas9 proteins. The fullength dCas9-G9a protein could be detected at the expected size of approximately 185kDa.
Figure 1: HA-tag fused to different dCas9-fusion proteins encoded on the RFC25 pSB1C3 backbone. dCas9-G9a under a CMV promoter is expressed at the expected size (185 kDa).
Functional tests
Targeting of dCas9-G9a to an open, endogenous locus should lead to changes in the epigenetic chromatin state and repression of gene expression. To test this, the endogenous VEGF-A locus was chosen, as it is well characterized and open in HEK-293T cells. Additionally, VEGF production can easily be measured by ELISA analysis.
Cells were seeded into 24-well format at a density of 65,000 cells per well and were co-transfected with the desired crRNAs, based on the RNAimer. These plasmids are derivates of the RNAimer, the only difference was the insertion of the target sites. These were: -8, -573, +434, -475 and -573/+434. As an off-target control crRNA targeting EMX was used. Every site was targeted with CMV:dCas9 wthout G9a, to differentiate the action of G9a from the CRISPRi effect. As an internal standard a constitutive SEAP reporter was used. 24 hours post transfection, VEGF levels and SEAP levels were measured and the ratio was calculated. Error bars represent standard deviations.
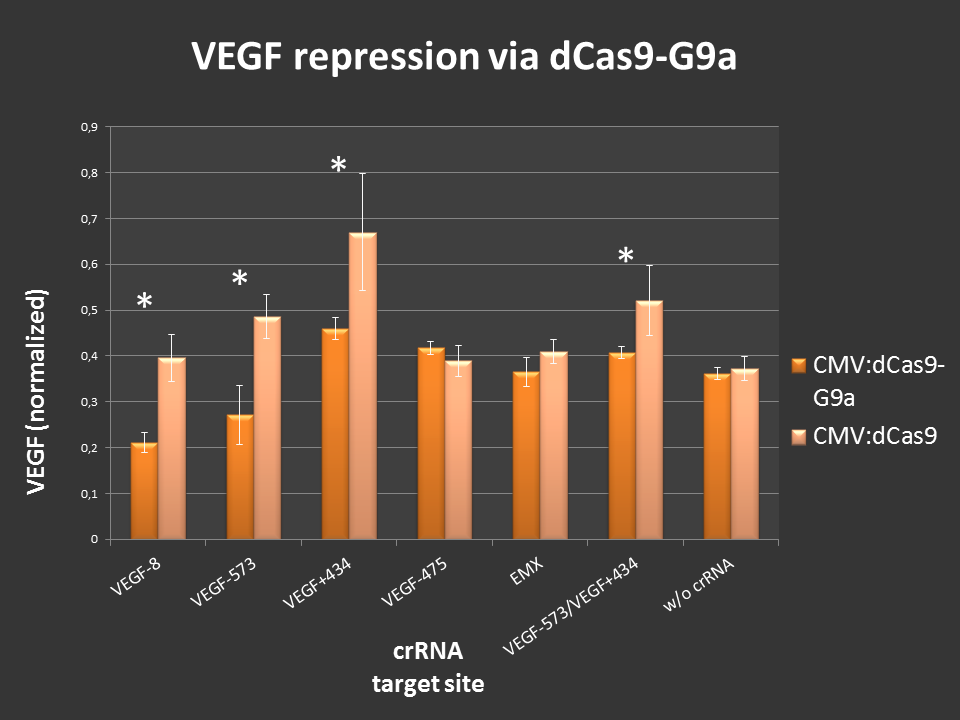
Figure 2: dCas9-G9a targeting of endogenous VEGF-A locus in HEK-293T cells.
ELISA values, normalized to internal SEAP standard. For some loci clear repressive effects up to 50% were detectable. Controls did not display any off-target effects. Combining two target sites did not show stronger repression than targeting one site at a time.
Different VEGF target sites led to different repression efficiencies and simultaneous targeting of two sites did not further enhance the observed effects.
In summary, the CMV:dCas9-G9a is an effective repressor of endogenous gene expression, by modulating the chromatin structure of the targeted locus. This offers application in fundamental research as well as in medical sciences.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 597
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| Protein data table for BioBrick BBa_ automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein) ATGGCCGGCGAGAATGTA ... ATCAACACCACCGGT ORF from nucleotide position -8 to 846 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC 25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
| Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges | ||||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
| Alignments (obtained from PredictProtein.org) There were no alignments for this protein in the data base. The BLAST search was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| Predictions (obtained from PredictProtein.org) | ||||||||||||||||||||||||||||||||||||||||||||||
| There were no predictions for this protein in the data base. The prediction was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
References
[1] Tachibana, M., et al. (2001). SET Domain-containing Protein, G9a, Is a Novel Lysine-preferring Mammalian Histone Methyltransferase with Hyperactivity and Specific Selectivity to Lysines 9 and 27 of Histone H3. The Journal of Biological Chemistry, 276, 25309-25317.
[2] Epsztejn-Litman, S., et al. (2008). De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 15(11): 1176–1183.
[3] Rizzini L., Favory J.-J., Cloix C., Faggionato D., O'Hara A., Kaiserli E., Baumeister R., Schäfer E., Nagy F., Jenkins G. I., Ulm R. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332