Difference between revisions of "Featured Parts:Fluorescent proteins"
Macampbell (Talk | contribs) |
Macampbell (Talk | contribs) |
||
| Line 27: | Line 27: | ||
<br> | <br> | ||
<br> | <br> | ||
| − | If you want to compare differnet FP, a series of comparable [http://parts.mit.edu/wiki/index.php/Davidson_Part_Nomenclature plasmids were generated] to permit standardized FP. This work began with part [https://parts.igem.org/cgi/partsdb/view.cgi?part_id=4479 BBa_i6060] which had a reputation as an unreliable FP. As you can read from the i6060 page, some aspects of FP is still | + | If you want to compare differnet FP, a series of comparable [http://parts.mit.edu/wiki/index.php/Davidson_Part_Nomenclature plasmids were generated] to permit standardized FP. This work began with part [https://parts.igem.org/cgi/partsdb/view.cgi?part_id=4479 BBa_i6060] which had a reputation as an unreliable FP. As you can read from the i6060 page, some aspects of FP is still not well documented. |
Revision as of 04:00, 8 April 2006

Fluorescent proteins (FP) are convenient was to visualize or quantify the output of a device or part. Many different FP have been cloned for use in the Registry.
The first FP to be cloned, was green fluorescent protein (GFP) which as its name implies gives its biological origin (a jellyfish) its green glow. From this starting place, several investigators generated mutations that altered the spectral properties of the FP. Now we have enhanced yellow fluorescent protein (EYFP), cyan fluorescent protein (CFP) and GFP.
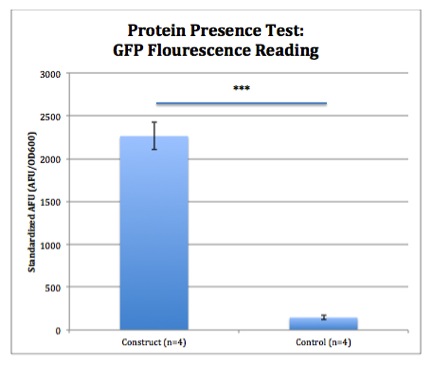
GFP visualized under UV light.
Red fluorescent protein (RFP) was isolated from a very different species (a coral) and its sequence is very different from the GFP family of FP. Several derivatives of RFP have been generated, most notably mOrange and mCherry.
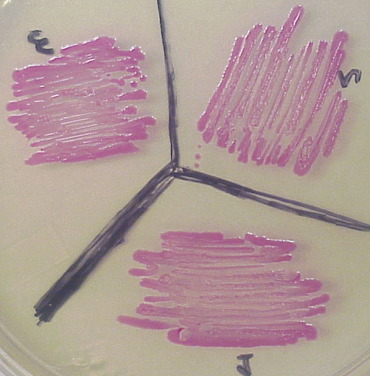
RFP visualized in white light. Note the colonies appear red at all times and fluoresce red as well.
One more feature you should know about are degradation tags; LVA and AAV. These short amino acid tags (~11 amino acids) are added to the carboxyl terminus to increase the rate of protein degradation and thus facilitate a shorter half-life of the tagged protein. These degradation tags work in prokaryotes and allow you to “turn off” a FP in favor of another reporter state. This was used in the classic represillator of Elowitz and Leibler (2000).
If you want to compare differnet FP, a series of comparable [http://parts.mit.edu/wiki/index.php/Davidson_Part_Nomenclature plasmids were generated] to permit standardized FP. This work began with part BBa_i6060 which had a reputation as an unreliable FP. As you can read from the i6060 page, some aspects of FP is still not well documented.
More Information/References
[http://www.bio.davidson.edu/courses/Molbio/restricted/02GFPseq/GFPseq.html Prashera, et al. 1992. Primary structure of the Aequorea victoria green-fluorescent protein. Gene, 111: 229-233.]
[http://www.bio.davidson.edu/courses/genomics/method/GFP.html Web page about GFP with interactive Jmol image.]
[http://www.bio.davidson.edu/courses/Molbio/restricted/02GFPwow/GFPwowpg1.html Chalfie, et al. 1994. Green Fluorescent Protein as a Marker for Gene Expression. Science. Vol. 263.]
[http://www.nature.com/cgi-taf/DynaPage.taf?file=/nature/journal/v403/n6767/abs/403335a0_fs.html&dynoptions=doi1043774228 Elowitz and Leibler. 2000. A synthetic oscillatory network of transcriptional regulators. Nature. 403: 335 - 338.]
[http://www.pubmedcentral.gov/articlerender.fcgi?tool=pubmed&pubmedid=11050229 Baird, et al. 2000. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. PNAS. 97(22): 11984–11989.]
